LIGHT controls distinct homeostatic and inflammatory gene expression profiles in esophageal fibroblasts via differential HVEM and LTβR-mediated mechanisms
- PMID: 34903876
- PMCID: PMC8866113
- DOI: 10.1038/s41385-021-00472-w
LIGHT controls distinct homeostatic and inflammatory gene expression profiles in esophageal fibroblasts via differential HVEM and LTβR-mediated mechanisms
Abstract
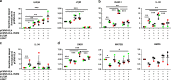
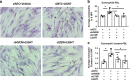
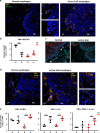
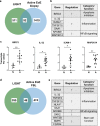
Fibroblasts mediate tissue remodeling in eosinophilic esophagitis (EoE), a chronic allergen-driven inflammatory pathology. Diverse fibroblast subtypes with homeostasis-regulating or inflammatory profiles have been recognized in various tissues, but which mediators induce these alternate differentiation states remain largely unknown. We recently identified that TNFSF14/LIGHT promotes an inflammatory esophageal fibroblast in vitro. Herein we used esophageal biopsies and primary fibroblasts to investigate the role of the LIGHT receptors, herpes virus entry mediator (HVEM) and lymphotoxin-beta receptor (LTβR), and their downstream activated pathways, in EoE. In addition to promoting inflammatory gene expression, LIGHT down-regulated homeostatic factors including WNTs, BMPs and type 3 semaphorins. In vivo, WNT2B+ fibroblasts were decreased while ICAM-1+ and IL-34+ fibroblasts were expanded in EoE, suggesting that a LIGHT-driven gene signature was imprinted in EoE versus normal esophageal fibroblasts. HVEM and LTβR overexpression and deficiency experiments demonstrated that HVEM regulates a limited subset of LIGHT targets, whereas LTβR controls all transcriptional effects. Pharmacologic blockade of the non-canonical NIK/p100/p52-mediated NF-κB pathway potently silenced LIGHT's transcriptional effects, with a lesser role found for p65 canonical NF-κB. Collectively, our results show that LIGHT promotes differentiation of esophageal fibroblasts toward an inflammatory phenotype and represses homeostatic gene expression via a LTβR-NIK-p52 NF-κB dominant pathway.
© 2021. The Author(s).
Conflict of interest statement
S.S.A. is a co-inventor of oral viscous budesonide for EoE patented by UCSD and licensed by Shire-Takeda. S.S.A. is a consultant for Regeneron, AImmune, Astellas, AstraZeneca, and Gossamer Bio. M.C has patents on TNFSF14/LIGHT. The remaining authors declare no competing interests.
Figures


References
-
- Manresa MC, et al. Hydroxylase inhibition regulates inflammation-induced intestinal fibrosis through the suppression of ERK-mediated TGF-beta 1 signaling (Vol 311, Pg G1076, 2016) Am. J. Physiol. Gastr L. 2017;312:G405–G405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

