Peroxisomal β-oxidation acts as a sensor for intracellular fatty acids and regulates lipolysis
- PMID: 34903883
- PMCID: PMC8688145
- DOI: 10.1038/s42255-021-00489-2
Peroxisomal β-oxidation acts as a sensor for intracellular fatty acids and regulates lipolysis
Abstract
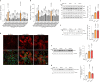
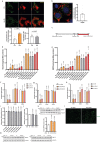
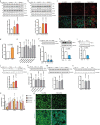
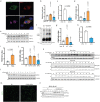
To liberate fatty acids (FAs) from intracellular stores, lipolysis is regulated by the activity of the lipases adipose triglyceride lipase (ATGL), hormone-sensitive lipase and monoacylglycerol lipase. Excessive FA release as a result of uncontrolled lipolysis results in lipotoxicity, which can in turn promote the progression of metabolic disorders. However, whether cells can directly sense FAs to maintain cellular lipid homeostasis is unknown. Here we report a sensing mechanism for cellular FAs based on peroxisomal degradation of FAs and coupled with reactive oxygen species (ROS) production, which in turn regulates FA release by modulating lipolysis. Changes in ROS levels are sensed by PEX2, which modulates ATGL levels through post-translational ubiquitination. We demonstrate the importance of this pathway for non-alcoholic fatty liver disease progression using genetic and pharmacological approaches to alter ROS levels in vivo, which can be utilized to increase hepatic ATGL levels and ameliorate hepatic steatosis. The discovery of this peroxisomal β-oxidation-mediated feedback mechanism, which is conserved in multiple organs, couples the functions of peroxisomes and lipid droplets and might serve as a new way to manipulate lipolysis to treat metabolic disorders.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

