Molecular dissection of a dedicated formaldehyde dehydrogenase from Mycobacterium smegmatis
- PMID: 34904319
- PMCID: PMC8862421
- DOI: 10.1002/pro.4258
Molecular dissection of a dedicated formaldehyde dehydrogenase from Mycobacterium smegmatis
Abstract
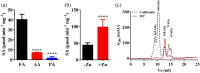
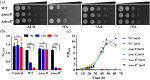
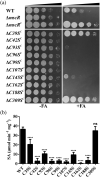
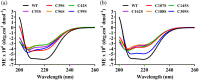
Accumulation of formaldehyde, a highly reactive molecule, in the cell is toxic, and requires detoxification for the organism's survival. Mycothiol-dependent formaldehyde dehydrogenase or S-nitrosomycothiol reductase (MscR) from Mycobacterium smegmatis and Mycobacterium tuberculosis was previously known for detoxifying formaldehyde and protecting the cell against nitrosative stress. We here show that M. smegmatis MscR exhibits a mycothiol-independent formaldehyde dehydrogenase (FDH) activity in vitro. Presence of zinc in the reaction enhances MscR activity, thus making it a zinc-dependent FDH. Interestingly, MscR utilizes only formaldehyde and no other primary aldehydes as its substrate in vitro, and M. smegmatis lacking mscR (ΔmscR) shows sensitivity exclusively toward formaldehyde. Bioinformatics analysis of MscRs from various bacteria reveals 10 positionally conserved cysteines, whose importance in structural stability and biological activity is not yet investigated. To explore the significance of these cysteines, we generated MscR single Cys variants by systematically replacing each cysteine with serine. All of the Cys variants except C39S and C309S are unable to show a complete rescue of ΔmscR on formaldehyde, show a significant loss of enzymatic activity in vitro, pronounced structural alterations as probed by circular dichroism, and loss of homotetramerization on size exclusion chromatography. Our data thus reveal the importance of intact cysteines in the structural stability and biological activity of MscR, which is a dedicated FDH in M. smegmatis, and shows ~84% identity with M. tuberculosis MscR. We believe that this knowledge will further help in the development of FDH as a potential drug target against M. tuberculosis infections.
Keywords: M. smegmatis; conserved cysteine; cysteine mutants; formaldehyde stress; mycothiol-dependent formaldehyde dehydrogenase; site directed mutagenesis.
© 2021 The Protein Society.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures


Similar articles
-
Deciphering the molecular mechanism and regulation of formaldehyde detoxification in Mycobacterium smegmatis.Appl Environ Microbiol. 2024 Feb 21;90(2):e0203923. doi: 10.1128/aem.02039-23. Epub 2024 Jan 23. Appl Environ Microbiol. 2024. PMID: 38259108 Free PMC article.
-
S-nitrosomycothiol reductase and mycothiol are required for survival under aldehyde stress and biofilm formation in Mycobacterium smegmatis.IUBMB Life. 2016 Aug;68(8):621-8. doi: 10.1002/iub.1524. Epub 2016 Jun 19. IUBMB Life. 2016. PMID: 27321674 Free PMC article.
-
Methylotrophy in Mycobacteria: Dissection of the Methanol Metabolism Pathway in Mycobacterium smegmatis.J Bacteriol. 2018 Aug 10;200(17):e00288-18. doi: 10.1128/JB.00288-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29891642 Free PMC article.
-
Mycothiol biochemistry.Arch Microbiol. 2002 Dec;178(6):388-94. doi: 10.1007/s00203-002-0469-4. Epub 2002 Sep 3. Arch Microbiol. 2002. PMID: 12420157 Review.
-
Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria.Microbiol Mol Biol Rev. 2008 Sep;72(3):471-94. doi: 10.1128/MMBR.00008-08. Microbiol Mol Biol Rev. 2008. PMID: 18772286 Free PMC article. Review.
Cited by
-
Elucidating the role of mycobacteriophage D29-encoded Gp36 in DNA binding and phage gene expression regulation.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf662. doi: 10.1093/nar/gkaf662. Nucleic Acids Res. 2025. PMID: 40671521 Free PMC article.
-
Deciphering the molecular mechanism and regulation of formaldehyde detoxification in Mycobacterium smegmatis.Appl Environ Microbiol. 2024 Feb 21;90(2):e0203923. doi: 10.1128/aem.02039-23. Epub 2024 Jan 23. Appl Environ Microbiol. 2024. PMID: 38259108 Free PMC article.
-
Photoradical-Mediated Catalyst-Independent Protein Cross-Link with Unusual Fluorescence Properties.Chembiochem. 2023 Sep 1;24(17):e202300380. doi: 10.1002/cbic.202300380. Epub 2023 Aug 1. Chembiochem. 2023. PMID: 37232210 Free PMC article.
-
Identification and characterization of a novel formaldehyde dehydrogenase in Bacillus subtilis.Appl Environ Microbiol. 2024 Nov 20;90(11):e0218123. doi: 10.1128/aem.02181-23. Epub 2024 Oct 29. Appl Environ Microbiol. 2024. PMID: 39470218 Free PMC article.
-
Competitive inhibition of a non-natural cofactor dependent formaldehyde dehydrogenase by imidazole.Biotechnol Lett. 2023 Jun;45(5-6):679-687. doi: 10.1007/s10529-023-03372-0. Epub 2023 Apr 18. Biotechnol Lett. 2023. PMID: 37071383
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

