An epigenetic switch activates bacterial quorum sensing and horizontal transfer of an integrative and conjugative element
- PMID: 34904658
- PMCID: PMC8789080
- DOI: 10.1093/nar/gkab1217
An epigenetic switch activates bacterial quorum sensing and horizontal transfer of an integrative and conjugative element
Abstract
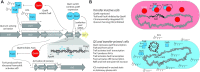
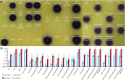
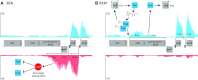
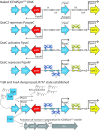
Horizontal transfer of the integrative and conjugative element ICEMlSymR7A converts non-symbiotic Mesorhizobium spp. into nitrogen-fixing legume symbionts. Here, we discover subpopulations of Mesorhizobium japonicum R7A become epigenetically primed for quorum-sensing (QS) and QS-activated horizontal transfer. Isolated populations in this state termed R7A* maintained these phenotypes in laboratory culture but did not transfer the R7A* state to recipients of ICEMlSymR7A following conjugation. We previously demonstrated ICEMlSymR7A transfer and QS are repressed by the antiactivator QseM in R7A populations and that the adjacently-coded DNA-binding protein QseC represses qseM transcription. Here RNA-sequencing revealed qseM expression was repressed in R7A* cells and that RNA antisense to qseC was abundant in R7A but not R7A*. Deletion of the antisense-qseC promoter converted cells into an R7A*-like state. An adjacently coded QseC2 protein bound two operator sites and repressed antisense-qseC transcription. Plasmid overexpression of QseC2 stimulated the R7A* state, which persisted following curing of this plasmid. The epigenetic maintenance of the R7A* state required ICEMlSymR7A-encoded copies of both qseC and qseC2. Therefore, QseC and QseC2, together with their DNA-binding sites and overlapping promoters, form a stable epigenetic switch that establishes binary control over qseM transcription and primes a subpopulation of R7A cells for QS and horizontal transfer.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Brockhurst M.A., Harrison E., Hall J.P.J., Richards T., McNally A., MacLean C.. The ecology and evolution of pangenomes. Curr. Biol. 2019; 29:R1094–R1103. - PubMed
-
- Hall J.P.J., Brockhurst M.A., Dytham C., Harrison E.. The evolution of plasmid stability: Are infectious transmission and compensatory evolution competing evolutionary trajectories?. Plasmid. 2017; 91:90–95. - PubMed
-
- Danino V.E., Wilkinson A., Edwards A., Downie J.A.. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 2003; 50:511–525. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

