pH/Acetonitrile-Gradient Reversed-Phase Fractionation of Enriched Hyper-Citrullinated Library in Combination with LC-MS/MS Analysis for Confident Identification of Citrullinated Peptides
- PMID: 34905169
- PMCID: PMC11552098
- DOI: 10.1007/978-1-0716-1936-0_9
pH/Acetonitrile-Gradient Reversed-Phase Fractionation of Enriched Hyper-Citrullinated Library in Combination with LC-MS/MS Analysis for Confident Identification of Citrullinated Peptides
Abstract
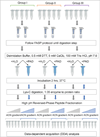
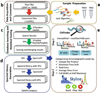
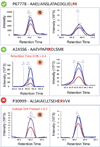
Citrullination, the Ca2+-driven enzymatic conversion of arginine residues to citrulline, is a posttranslational modification, implicated in several physiological and pathological processes. Several methods to detect citrullinated proteins have been developed, including color development reagent, fluorescence, phenylglyoxal, and antibody-based methods. These methods yet suffer from limitations in sensitivity, specificity, or citrullinated site determination. Mass spectrometry (MS)-based proteomic analysis has emerged as a promising method to resolve these problems. However, due to low abundance of citrullinated proteins and similar MS features to deamidation of asparagine and glutamine, confident identification of citrullinated proteome is challenging. Here, we present a systematic approach to identify a compendium of steps to enhance the number of detected citrullinated residue and implement diagnostic MS feature that allow the confidence of MS-based identifications. Our method is based on the concept of generation of hyper-citrullinated library with high-pH reversed-phase peptide fractionation that allows to enrich in low abundance citrullinated peptides and amplify the effect of charge loss upon citrullination. Application of our approach to complex global citrullino-proteome datasets demonstrates the confident assessment of citrullinated peptides, thereby enhancing the size and functional interpretation of citrullinated proteomes.
Keywords: Citrullination; Deamidation; Hyper-citrullinated peptide library; Mass spectrometry; Retention time; pH reversed-phase peptide fractionation.
© 2022. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures

Similar articles
-
Statistical Modeling for Enhancing the Discovery Power of Citrullination from Tandem Mass Spectrometry Data.Anal Chem. 2020 Oct 6;92(19):12975-12986. doi: 10.1021/acs.analchem.0c01687. Epub 2020 Sep 16. Anal Chem. 2020. PMID: 32876429
-
Confident Identification of Citrullination and Carbamylation Assisted by Peptide Retention Time Prediction.J Proteome Res. 2021 Mar 5;20(3):1571-1581. doi: 10.1021/acs.jproteome.0c00775. Epub 2021 Feb 1. J Proteome Res. 2021. PMID: 33523662
-
Fractionation of Enriched Phosphopeptides Using pH/Acetonitrile-Gradient-Reversed-Phase Microcolumn Separation in Combination with LC-MS/MS Analysis.Int J Mol Sci. 2020 Jun 1;21(11):3971. doi: 10.3390/ijms21113971. Int J Mol Sci. 2020. PMID: 32492839 Free PMC article.
-
[Advances in high-throughput proteomic analysis].Se Pu. 2021 Feb;39(2):112-117. doi: 10.3724/SP.J.1123.2020.08023. Se Pu. 2021. PMID: 34227342 Free PMC article. Review. Chinese.
-
Characterizing citrullination by mass spectrometry-based proteomics.Philos Trans R Soc Lond B Biol Sci. 2023 Nov 20;378(1890):20220237. doi: 10.1098/rstb.2022.0237. Epub 2023 Oct 2. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 37778389 Free PMC article. Review.
Cited by
-
Protein arginine deiminase 2 (PAD2) modulates the polarization of THP-1 macrophages to the anti-inflammatory M2 phenotype.J Inflamm (Lond). 2022 Nov 18;19(1):20. doi: 10.1186/s12950-022-00317-8. J Inflamm (Lond). 2022. PMID: 36401289 Free PMC article.
-
The Future of Proteomics is Up in the Air: Can Ion Mobility Replace Liquid Chromatography for High Throughput Proteomics?J Proteome Res. 2024 Jun 7;23(6):1871-1882. doi: 10.1021/acs.jproteome.4c00248. Epub 2024 May 7. J Proteome Res. 2024. PMID: 38713528 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

