Clinico-histopathologic and single-nuclei RNA-sequencing insights into cardiac injury and microthrombi in critical COVID-19
- PMID: 34905515
- PMCID: PMC8855793
- DOI: 10.1172/jci.insight.154633
Clinico-histopathologic and single-nuclei RNA-sequencing insights into cardiac injury and microthrombi in critical COVID-19
Abstract
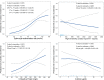
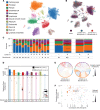
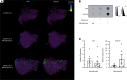
Acute cardiac injury is prevalent in critical COVID-19 and associated with increased mortality. Its etiology remains debated, as initially presumed causes - myocarditis and cardiac necrosis - have proved uncommon. To elucidate the pathophysiology of COVID-19-associated cardiac injury, we conducted a prospective study of the first 69 consecutive COVID-19 decedents at CUIMC in New York City. Of 6 acute cardiac histopathologic features, presence of microthrombi was the most commonly detected among our cohort. We tested associations of cardiac microthrombi with biomarkers of inflammation, cardiac injury, and fibrinolysis and with in-hospital antiplatelet therapy, therapeutic anticoagulation, and corticosteroid treatment, while adjusting for multiple clinical factors, including COVID-19 therapies. Higher peak erythrocyte sedimentation rate and C-reactive protein were independently associated with increased odds of microthrombi, supporting an immunothrombotic etiology. Using single-nuclei RNA-sequencing analysis on 3 patients with and 4 patients without cardiac microthrombi, we discovered an enrichment of prothrombotic/antifibrinolytic, extracellular matrix remodeling, and immune-potentiating signaling among cardiac fibroblasts in microthrombi-positive, relative to microthrombi-negative, COVID-19 hearts. Non-COVID-19, nonfailing hearts were used as reference controls. Our study identifies a specific transcriptomic signature in cardiac fibroblasts as a salient feature of microthrombi-positive COVID-19 hearts. Our findings warrant further mechanistic study as cardiac fibroblasts may represent a potential therapeutic target for COVID-19-associated cardiac microthrombi.
Keywords: Bioinformatics; COVID-19; Cardiology; Cardiovascular disease; Molecular pathology.
Figures

Update of
-
Molecular Pathophysiology of Cardiac Injury and Cardiac Microthrombi in Fatal COVID-19: Insights from Clinico-histopathologic and Single Nuclei RNA Sequencing Analyses.bioRxiv [Preprint]. 2021 Jul 27:2021.07.27.453843. doi: 10.1101/2021.07.27.453843. bioRxiv. 2021. Update in: JCI Insight. 2022 Jan 25;7(2):e154633. doi: 10.1172/jci.insight.154633. PMID: 34341789 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

