Quantifying prediction of pathogenicity for within-codon concordance (PM5) using 7541 functional classifications of BRCA1 and MSH2 missense variants
- PMID: 34906453
- PMCID: PMC8896276
- DOI: 10.1016/j.gim.2021.11.011
Quantifying prediction of pathogenicity for within-codon concordance (PM5) using 7541 functional classifications of BRCA1 and MSH2 missense variants
Abstract
Purpose: Conditions and thresholds applied for evidence weighting of within-codon concordance (PM5) for pathogenicity vary widely between laboratories and expert groups. Because of the sparseness of available clinical classifications, there is little evidence for variation in practice.
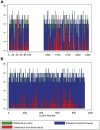
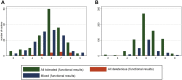
Methods: We used as a truthset 7541 dichotomous functional classifications of BRCA1 and MSH2, spanning 311 codons of BRCA1 and 918 codons of MSH2, generated from large-scale functional assays that have been shown to correlate excellently with clinical classifications. We assessed PM5 at 5 stringencies with incorporation of 8 in silico tools. For each analysis, we quantified a positive likelihood ratio (pLR, true positive rate/false positive rate), the predictive value of PM5-lookup in ClinVar compared with the functional truthset.
Results: pLR was 16.3 (10.6-24.9) for variants for which there was exactly 1 additional colocated deleterious variant on ClinVar, and the variant under examination was equally or more damaging when analyzed using BLOSUM62. pLR was 71.5 (37.8-135.3) for variants for which there were 2 or more colocated deleterious ClinVar variants, and the variant under examination was equally or more damaging than at least 1 colocated variant when analyzed using BLOSUM62.
Conclusion: These analyses support the graded use of PM5, with potential to use it at higher evidence weighting where more stringent criteria are met.
Keywords: ACMG; Classification; Codon; PM5; Variant.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures

References
-
- Firth H.V., Hurst J.A. 2nd ed. Oxford University Press; 2017. Oxford Desk Reference: Clinical Genetics and Genomics.
-
- Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- Rivera-Muñoz E.A., Milko L.V., Harrison S.M., et al. ClinGen Variant Curation Expert Panel experiences and standardized processes for disease and gene-level specification of the ACMG/AMP guidelines for sequence variant interpretation. Hum Mutat. 2018;39(11):1614–1622. doi: 10.1002/humu.23645. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

