Recognition of G-quadruplex RNA by a crucial RNA methyltransferase component, METTL14
- PMID: 34908152
- PMCID: PMC8755082
- DOI: 10.1093/nar/gkab1211
Recognition of G-quadruplex RNA by a crucial RNA methyltransferase component, METTL14
Abstract
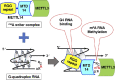
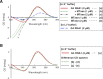
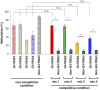
N6-methyladenosine (m6A) is an important epitranscriptomic chemical modification that is mainly catalyzed by the METTL3/METTL14 RNA methyltransferase heterodimer. Although m6A is found at the consensus sequence of 5'-DRACH-3' in various transcripts, the mechanism by which METTL3/METTL14 determines its target is unclear. This study aimed to clarify the RNA binding property of METTL3/METTL14. We found that the methyltransferase heterodimer itself has a binding preference for RNA G-quadruplex (rG4) structures, which are non-canonical four-stranded structures formed by G-rich sequences, via the METTL14 RGG repeats. Additionally, the methyltransferase heterodimer selectively methylated adenosines close to the rG4 sequences. These results suggest a possible process for direct recruitment of METTL3/METTL14 to specific methylation sites, especially near the G4-forming regions. This study is the first to report the RNA binding preference of the m6A writer complex for the rG4 structure and provides insights into the role of rG4 in epitranscriptomic regulation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures