Macroscopic Structural and Connectome Mapping of the Mouse Brain Using Diffusion Magnetic Resonance Imaging
- PMID: 34909442
- PMCID: PMC8635841
- DOI: 10.21769/BioProtoc.4221
Macroscopic Structural and Connectome Mapping of the Mouse Brain Using Diffusion Magnetic Resonance Imaging
Abstract
Translational work in rodents elucidates basic mechanisms that drive complex behaviors relevant to psychiatric and neurological conditions. Nonetheless, numerous promising studies in rodents later fail in clinical trials, highlighting the need for improving the translational utility of preclinical studies in rodents. Imaging of small rodents provides an important strategy to address this challenge, as it enables a whole-brain unbiased search for structural and dynamic changes that can be directly compared to human imaging. The functional significance of structural changes identified using imaging can then be further investigated using molecular and genetic tools available for the mouse. Here, we describe a pipeline for unbiased search and characterization of structural changes and network properties, based on diffusion MRI data covering the entire mouse brain at an isotropic resolution of 100 µm. We first used unbiased whole-brain voxel-based analyses to identify volumetric and microstructural alterations in the brain of adult mice exposed to unpredictable postnatal stress (UPS), which is a mouse model of complex early life stress (ELS). Brain regions showing structural abnormalities were used as nodes to generate a grid for assessing structural connectivity and network properties based on graph theory. The technique described here can be broadly applied to understand brain connectivity in other mouse models of human disorders, as well as in genetically modified mouse strains. Graphic abstract: Pipeline for characterizing structural connectome in the mouse brain using diffusion magnetic resonance imaging. Scale bar = 1 mm.
Keywords: Brain network properties; Diffusion MRI; Fiber tractography; Mouse brain.; Structural connectivity.
©Copyright Arefin et al.
Conflict of interest statement
Competing interestsThe authors declare no conflict of interest.
Figures

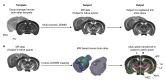


References
-
- Dorr A. E., Lerch J. P., Spring S., Kabani N. and Henkelman R. M.(2008). High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage 42(1):60-9. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

