This is a preprint.
Insights on the mutational landscape of the SARS-CoV-2 Omicron variant
- PMID: 34909771
- PMCID: PMC8669838
- DOI: 10.1101/2021.12.06.471499
Insights on the mutational landscape of the SARS-CoV-2 Omicron variant
Update in
-
Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain.Cell Rep Med. 2022 Jan 24;3(2):100527. doi: 10.1016/j.xcrm.2022.100527. eCollection 2022 Feb 15. Cell Rep Med. 2022. PMID: 35233548 Free PMC article.
Abstract
The SARS-COV2 Omicron variant has sparked global concern due to the possibility of enhanced transmissibility and escape from vaccines and therapeutics. In this study, we describe the mutational landscape of the Omicron variant using amino acid interaction (AAI) networks. AAI network analysis is particularly well suited for interrogating the impact of constellations of mutations as occur on Omicron that may function in an epistatic manner. Our analyses suggest that as compared to previous variants of concern, the Omicron variant has increased antibody escape breadth due to mutations in class 3 and 4 antibody epitopes as well as increased escape depth due to accumulated mutations in class 1 antibody epitopes. We note certain RBD mutations that might further enhance Omicron's escape, and in particular advise careful surveillance of two subclades bearing R346S/K mutations with relevance for certain therapeutic antibodies. Further, AAI network analysis suggests that the function of certain therapeutic monoclonal antibodies may be disrupted by Omicron mutations as a result of the cumulative indirect perturbations to the epitope surface properties, despite point-mutation analyses suggesting these antibodies are tolerant of the set of Omicron mutations in isolation. Finally, for several Omicron mutations that do not appear to contribute meaningfully to antibody escape, we find evidence for a plausible role in enhanced transmissibility via disruption of RBD-down conformational stability at the RBD-RBD interface.
Keywords: Antibody Escape; Antigenic Drift; Epistasis; Mutation; Network Analysis; Omicron; RBD Interface Stability; SARS-CoV-2; Variant of Concern.
Conflict of interest statement
Competing Interests RS is a board member of Tychan Pte. Ltd Singapore, which focuses on Infectious Diseases.
Figures
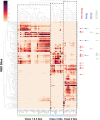
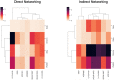

References
-
- Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020. Dec;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. Epub 2020 Oct 12. - DOI - PMC - PubMed
-
- Cathcart AL, Havenar-Daughton C, Lempp FA, et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv 2021.03.09.434607; doi: 10.1101/2021.03.09.434607 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
