Improvement of putrescine production through the arginine decarboxylase pathway in Escherichia coli K-12
- PMID: 34910273
- PMCID: PMC8674398
- DOI: 10.1186/s13568-021-01330-5
Improvement of putrescine production through the arginine decarboxylase pathway in Escherichia coli K-12
Abstract
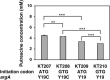
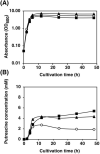
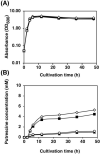
In the bio-based polymer industry, putrescine is in the spotlight for use as a material. We constructed strains of Escherichia coli to assess its putrescine production capabilities through the arginine decarboxylase pathway in batch fermentation. N-Acetylglutamate (ArgA) synthase is subjected to feedback inhibition by arginine. Therefore, the 19th amino acid residue, Tyr, of argA was substituted with Cys to desensitize the feedback inhibition of arginine, resulting in improved putrescine production. The inefficient initiation codon GTG of argA was substituted with the effective ATG codon, but its replacement did not affect putrescine production. The essential genes for the putrescine production pathway, speA and speB, were cloned into the same plasmid with argAATG Y19C to form an operon. These genes were introduced under different promoters; lacIp, lacIqp, lacIq1p, and T5p. Among these, the T5 promoter demonstrated the best putrescine production. In addition, disruption of the puuA gene encoding enzyme of the first step of putrescine degradation pathway increased the putrescine production. Of note, putrescine production was not affected by the disruption of patA, which encodes putrescine aminotransferase, the initial enzyme of another putrescine utilization pathway. We also report that the strain KT160, which has a genomic mutation of YifEQ100TAG, had the greatest putrescine production. At 48 h of batch fermentation, strain KT160 grown in terrific broth with 0.01 mM IPTG produced 19.8 mM of putrescine.
Keywords: Glutamate-putrescine ligase; Macrodomain Ori protein; N-acetylglutamate synthase; Terrific broth.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures





Similar articles
-
Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains.Appl Environ Microbiol. 1998 May;64(5):1805-11. doi: 10.1128/AEM.64.5.1805-1811.1998. Appl Environ Microbiol. 1998. PMID: 9572954 Free PMC article.
-
A novel strategy for L-arginine production in engineered Escherichia coli.Microb Cell Fact. 2023 Jul 26;22(1):138. doi: 10.1186/s12934-023-02145-8. Microb Cell Fact. 2023. PMID: 37495979 Free PMC article.
-
Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system.Appl Microbiol Biotechnol. 2012 Jul;95(1):169-78. doi: 10.1007/s00253-012-3956-9. Epub 2012 Feb 28. Appl Microbiol Biotechnol. 2012. PMID: 22370950
-
Cyclic AMP inhibits and putrescine represses expression of the speA gene encoding biosynthetic arginine decarboxylase in Escherichia coli.J Bacteriol. 1991 Jun;173(12):3615-21. doi: 10.1128/jb.173.12.3615-3621.1991. J Bacteriol. 1991. PMID: 1646785 Free PMC article.
-
gamma-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12.J Biol Chem. 2008 Jul 18;283(29):19981-90. doi: 10.1074/jbc.M800133200. Epub 2008 May 21. J Biol Chem. 2008. PMID: 18495664
Cited by
-
Rhizobium biostimulation of blackberry modulates survival pathways in Caenorhabditis elegans across biological kingdoms.NPJ Sci Food. 2025 Jul 29;9(1):160. doi: 10.1038/s41538-025-00525-5. NPJ Sci Food. 2025. PMID: 40730576 Free PMC article.
-
Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum.Bioengineering (Basel). 2023 Jan 24;10(2):157. doi: 10.3390/bioengineering10020157. Bioengineering (Basel). 2023. PMID: 36829651 Free PMC article.
-
Putrescine Supplementation Limits the Expansion of pks+ Escherichia coli and Tumor Development in the Colon.Cancer Res Commun. 2024 Jul 1;4(7):1777-1792. doi: 10.1158/2767-9764.CRC-23-0355. Cancer Res Commun. 2024. PMID: 38934090 Free PMC article.
-
Antibiofilm and Antivirulence Properties of 6-Polyaminosteroid Derivatives against Antibiotic-Resistant Bacteria.Antibiotics (Basel). 2023 Dec 20;13(1):8. doi: 10.3390/antibiotics13010008. Antibiotics (Basel). 2023. PMID: 38275318 Free PMC article.
-
Polyamines: their significance for maintaining health and contributing to diseases.Cell Commun Signal. 2023 Dec 4;21(1):348. doi: 10.1186/s12964-023-01373-0. Cell Commun Signal. 2023. PMID: 38049863 Free PMC article. Review.
References
-
- Demco DE, Litvinov VM, Rata G, Popescu C, Phan KH, Schmidt A, Blümich B. Investigation of thermal aging of polyamide 4, 6 by 1H solid-state NMR. Macromol Chem Phys. 2007;208:2085–2095. doi: 10.1002/macp.200700095. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

