Isolation and Characterization of Levoglucosan-Metabolizing Bacteria
- PMID: 34910566
- PMCID: PMC8863068
- DOI: 10.1128/AEM.01868-21
Isolation and Characterization of Levoglucosan-Metabolizing Bacteria
Abstract
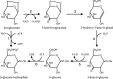
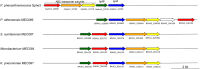
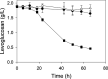
Bacteria were isolated from wastewater and soil containing charred wood remnants based on their ability to use levoglucosan as a sole carbon source and on their levoglucosan dehydrogenase (LGDH) activity. On the basis of their 16S rRNA gene sequences, these bacteria represented the diverse genera Microbacterium, Paenibacillus, Shinella, and Klebsiella. Genomic sequencing of the isolates verified that two isolates represented novel species, Paenibacillus athensensis MEC069T and Shinella sumterensis MEC087T, while the remaining isolates were closely related to Microbacterium lacusdiani or Klebsiella pneumoniae. The genetic sequence of LGDH, lgdA, was found in the genomes of these four isolates as well as Pseudarthrobacter phenanthrenivorans Sphe3. The identity of the P. phenanthrenivorans LGDH was experimentally verified following recombinant expression in Escherichia coli. Comparison of the putative genes surrounding lgdA in the isolate genomes indicated that several other gene products facilitate the bacterial catabolism of levoglucosan, including a putative sugar isomerase and several transport proteins. IMPORTANCE Levoglucosan is the most prevalent soluble carbohydrate remaining after high-temperature pyrolysis of lignocellulosic biomass, but it is not fermented by typical production microbes such as Escherichia coli and Saccharomyces cerevisiae. A few fungi metabolize levoglucosan via the enzyme levoglucosan kinase, while several bacteria metabolize levoglucosan via levoglucosan dehydrogenase. This study describes the isolation and characterization of four bacterial species that degrade levoglucosan. Each isolate is shown to contain several genes within an operon involved in levoglucosan degradation, furthering our understanding of bacteria that metabolize levoglucosan.
Keywords: Klebsiella; Microbacterium; Paenibacillus; Pseudarthrobacter phenanthrenivorans; Shinella; levoglucosan dehydrogenase; pyrolysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Maduskar S, Maliekkal V, Neurock M, Dauenhauer PJ. 2018. On the yield of levoglucosan from cellulose pyrolysis. ACS Sustainable Chem Eng 6:7017–7025. 10.1021/acssuschemeng.8b00853. - DOI
-
- Junior II, do Nascimento MA, Alves de Souza ROM, Dufour A, Wojcieszak R. 2020. Levoglucosan: a promising platform molecule? Green Chem 22:5859–5880. 10.1039/D0GC01490G. - DOI
-
- Lomax JA, Commandeur JM, Arisz PW, Boon JA. 1991. Characterization of oligomers and sugar ring-cleavage products in the pyrolysate of cellulose. J Anal App Pyrol 19:65–79. 10.1016/0165-2370(91)80035-7. - DOI
-
- Dobele G, Rossinskaja G, Dizhbite T, Telysheva G, Meier D, Faix O. 2005. Application of catalysis for obtaining 1,6-anhydrosaccharides from cellulose and wood by fast pyrolysis. J Anal Appl Pyrolysis 74:401–405. 10.1016/j.jaap.2004.11.031. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

