Comparison of Whole-Genome Sequence-Based Methods and PCR Ribotyping for Subtyping of Clostridioides difficile
- PMID: 34911367
- PMCID: PMC8849210
- DOI: 10.1128/JCM.01737-21
Comparison of Whole-Genome Sequence-Based Methods and PCR Ribotyping for Subtyping of Clostridioides difficile
Abstract
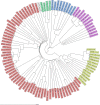
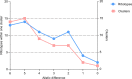
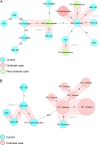
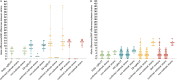
Clostridioides difficile is the most common cause of antibiotic-associated gastrointestinal infections. Capillary electrophoresis (CE)-PCR ribotyping is currently the gold standard for C. difficile typing but lacks the discriminatory power to study transmission and outbreaks in detail. New molecular methods have the capacity to differentiate better and provide standardized and interlaboratory exchangeable data. Using a well-characterized collection of diverse strains (N = 630; 100 unique ribotypes [RTs]), we compared the discriminatory power of core genome multilocus sequence typing (cgMLST) (SeqSphere and EnteroBase), whole-genome MLST (wgMLST) (EnteroBase), and single-nucleotide polymorphism (SNP) analysis. A unique cgMLST profile (more than six allele differences) was observed in 82 of 100 RTs, indicating that cgMLST could distinguish most, but not all, RTs. Application of cgMLST in two outbreak settings with RT078 and RT181 (known to have low intra-RT allele differences) showed no distinction between outbreak and nonoutbreak strains in contrast to wgMLST and SNP analysis. We conclude that cgMLST has the potential to be an alternative to CE-PCR ribotyping. The method is reproducible, easy to standardize, and offers higher discrimination. However, adjusted cutoff thresholds and epidemiological data are necessary to recognize outbreaks of some specific RTs. We propose to use an allelic threshold of three alleles to identify outbreaks.
Keywords: Clostridioides difficile; core-genome MLST; typing methods; whole-genome MLST; whole-genome sequencing.
Conflict of interest statement
The authors declare a conflict of interest. D.W.E.: lecture fees from Gilead, outside the submitted work; E.J.K.: unrestricted research grant from Vedanta Bioscience, Boston.
D.W.E. received lecture fees from Gilead, outside the submitted work. E.J.K. received an unrestricted research grant from Vedanta Bioscience (Boston, MA). The other authors declare no conflict of interest.
Figures

References
-
- Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57:1377–1382. 10.1099/jmm.0.47714-0. - DOI - PMC - PubMed
-
- Fawley WN, Knetsch CW, MacCannell DR, Harmanus C, Du T, Mulvey MR, Paulick A, Anderson L, Kuijper EJ, Wilcox MH. 2015. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One 10:e0118150. 10.1371/journal.pone.0118150. - DOI - PMC - PubMed
-
- Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 48:770–778. 10.1128/JCM.01796-09. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

