Protein phosphatase 2A inactivation induces microsatellite instability, neoantigen production and immune response
- PMID: 34911954
- PMCID: PMC8674339
- DOI: 10.1038/s41467-021-27620-x
Protein phosphatase 2A inactivation induces microsatellite instability, neoantigen production and immune response
Abstract
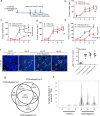
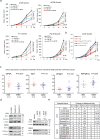
Microsatellite-instable (MSI), a predictive biomarker for immune checkpoint blockade (ICB) response, is caused by mismatch repair deficiency (MMRd) that occurs through genetic or epigenetic silencing of MMR genes. Here, we report a mechanism of MMRd and demonstrate that protein phosphatase 2A (PP2A) deletion or inactivation converts cold microsatellite-stable (MSS) into MSI tumours through two orthogonal pathways: (i) by increasing retinoblastoma protein phosphorylation that leads to E2F and DNMT3A/3B expression with subsequent DNA methylation, and (ii) by increasing histone deacetylase (HDAC)2 phosphorylation that subsequently decreases H3K9ac levels and histone acetylation, which induces epigenetic silencing of MLH1. In mouse models of MSS and MSI colorectal cancers, triple-negative breast cancer and pancreatic cancer, PP2A inhibition triggers neoantigen production, cytotoxic T cell infiltration and ICB sensitization. Human cancer cell lines and tissue array effectively confirm these signaling pathways. These data indicate the dual involvement of PP2A inactivation in silencing MLH1 and inducing MSI.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gryfe R, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342:69–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

