octoFLUshow: an Interactive Tool Describing Spatial and Temporal Trends in the Genetic Diversity of Influenza A Virus in U.S. Swine
- PMID: 34913720
- PMCID: PMC8675265
- DOI: 10.1128/MRA.01081-21
octoFLUshow: an Interactive Tool Describing Spatial and Temporal Trends in the Genetic Diversity of Influenza A Virus in U.S. Swine
Abstract
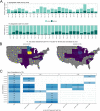
Influenza A virus (IAV) is passively surveilled in swine in the United States through a U.S. Department of Agriculture administered surveillance system. We present an interactive Web tool to visualize and explore trends in the genetic and geographic diversity of IAV derived from the surveillance system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rajao DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, Vincent AL. 2017. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 91:e01763-16. doi:10.1128/JVI.01763-16. - DOI - PMC - PubMed
Grants and funding
- DE-AC05-06OR23100/Oak Ridge Associated Universities (ORAU)
- 5030-32000-231-080-I/USDA | Animal and Plant Health Inspection Service (APHIS)
- 5030-32000-231-000D/USDA | Agricultural Research Service (ARS)
- 0500-00093-001-00-D/USDA | Agricultural Research Service (ARS)
- 75N93021C00015/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
