Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe
- PMID: 34914036
- PMCID: PMC8675301
- DOI: 10.1007/s15010-021-01734-2
Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe
Erratum in
-
Correction to: Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe.Infection. 2022 Apr;50(2):327. doi: 10.1007/s15010-021-01752-0. Infection. 2022. PMID: 35031979 Free PMC article. No abstract available.
Abstract
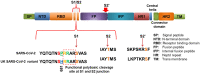
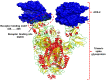
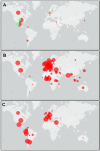
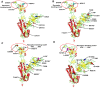
The coronavirus disease-19 has left a permanent mark on the history of the human race. Severe acute respiratory syndrome coronavirus-2 is a positive-sense single-stranded RNA virus, first reported in Wuhan, China, in December 2019 and from there took over the world. Being highly susceptible to mutations, the virus's numerous variants started to appear, and some were more lethal and infectious than the parent. The effectiveness of the vaccine is also affected severely against the new variant. In this study, the infectious mechanism of the coronavirus is explained with a focus on different variants and their respective mutations, which play a critical role in the increased transmissibility, infectivity, and immune escape of the virus. As India has already faced the second wave of the pandemic, the future outlook on the likeliness of a third wave with respect to the Indian variants such as kappa, delta, and Delta Plus is also discussed. This review article aims to reflect the catastrophe of the variants of SARS-CoV-2 and the possibility of developing even more severe variants in the near future.
Keywords: Immune evasion; Mutations; SARS-CoV-2; Vaccines; Variants; Waves.
© 2021. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



References
-
- WHO Coronavirus (COVID-19) Dashboard. WHO (2021). https://covid19.who.int/. Accessed 10 July 2021.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

