TIM3+ TRBV11-2 T cells and IFNγ signature in patrolling monocytes and CD16+ NK cells delineate MIS-C
- PMID: 34914824
- PMCID: PMC8685281
- DOI: 10.1084/jem.20211381
TIM3+ TRBV11-2 T cells and IFNγ signature in patrolling monocytes and CD16+ NK cells delineate MIS-C
Abstract
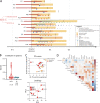
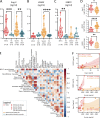
In rare instances, pediatric SARS-CoV-2 infection results in a novel immunodysregulation syndrome termed multisystem inflammatory syndrome in children (MIS-C). We compared MIS-C immunopathology with severe COVID-19 in adults. MIS-C does not result in pneumocyte damage but is associated with vascular endotheliitis and gastrointestinal epithelial injury. In MIS-C, the cytokine release syndrome is characterized by IFNγ and not type I interferon. Persistence of patrolling monocytes differentiates MIS-C from severe COVID-19, which is dominated by HLA-DRlo classical monocytes. IFNγ levels correlate with granzyme B production in CD16+ NK cells and TIM3 expression on CD38+/HLA-DR+ T cells. Single-cell TCR profiling reveals a skewed TCRβ repertoire enriched for TRBV11-2 and a superantigenic signature in TIM3+/CD38+/HLA-DR+ T cells. Using NicheNet, we confirm IFNγ as a central cytokine in the communication between TIM3+/CD38+/HLA-DR+ T cells, CD16+ NK cells, and patrolling monocytes. Normalization of IFNγ, loss of TIM3, quiescence of CD16+ NK cells, and contraction of patrolling monocytes upon clinical resolution highlight their potential role in MIS-C immunopathogenesis.
© 2021 Hoste et al.
Conflict of interest statement
Disclosures: No disclosures were reported.
Figures












References
-
- Abrams, J.Y., Oster M.E., Godfred-Cato S.E., Bryant B., Datta S.D., Campbell A.P., Leung J.W., Tsang C.A., Pierce T.J., Kennedy J.L., et al. 2021. Factors linked to severe outcomes in multisystem inflammatory syndrome in children (MIS-C) in the USA: a retrospective surveillance study. Lancet Child Adolesc. Health. 5:323–331. 10.1016/S2352-4642(21)00050-X - DOI - PMC - PubMed
-
- Bastard, P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. COVID Human Genetic Effort . 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370:eabd4585. 10.1126/science.abd4585 - DOI - PMC - PubMed
-
- Beckmann, N.D., Comella P.H., Cheng E., Lepow L., Beckmann A.G., Mouskas K., Simons N.W., Hoffman G.E., Francoeur N.J., Del Valle D.M., et al. 2020. Cytotoxic lymphocytes are dysregulated in multisystem inflammatory syndrome in children. medRxiv. 10.1101/2020.08.29.20182899 (Preprint posted September 2, 2020) - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

