R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability
- PMID: 34916496
- PMCID: PMC8677849
- DOI: 10.1038/s41467-021-27530-y
R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability
Abstract
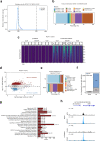
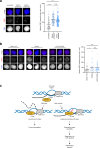
Transcription poses a threat to genomic stability through the formation of R-loops that can obstruct progression of replication forks. R-loops are three-stranded nucleic acid structures formed by an RNA-DNA hybrid with a displaced non-template DNA strand. We developed RNA-DNA Proximity Proteomics to map the R-loop proximal proteome of human cells using quantitative mass spectrometry. We implicate different cellular proteins in R-loop regulation and identify a role of the tumor suppressor DDX41 in opposing R-loop and double strand DNA break accumulation in promoters. DDX41 is enriched in promoter regions in vivo, and can unwind RNA-DNA hybrids in vitro. R-loop accumulation upon loss of DDX41 is accompanied with replication stress, an increase in the formation of double strand DNA breaks and transcriptome changes associated with the inflammatory response. Germline loss-of-function mutations in DDX41 lead to predisposition to acute myeloid leukemia in adulthood. We propose that R-loop accumulation and genomic instability-associated inflammatory response may contribute to the development of familial AML with mutated DDX41.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

