Induction of Rosette-to-Lumen stage embryoids using reprogramming paradigms in ESCs
- PMID: 34916498
- PMCID: PMC8677818
- DOI: 10.1038/s41467-021-27586-w
Induction of Rosette-to-Lumen stage embryoids using reprogramming paradigms in ESCs
Abstract
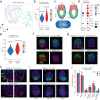
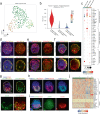
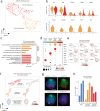
Blastocyst-derived stem cell lines were shown to self-organize into embryo-like structures in 3D cell culture environments. Here, we provide evidence that embryo-like structures can be generated solely based on transcription factor-mediated reprogramming of embryonic stem cells in a simple 3D co-culture system. Embryonic stem cells in these cultures self-organize into elongated, compartmentalized embryo-like structures reflecting aspects of the inner regions of the early post-implantation embryo. Single-cell RNA-sequencing reveals transcriptional profiles resembling epiblast, primitive-/visceral endoderm, and extraembryonic ectoderm of early murine embryos around E4.5-E5.5. In this stem cell-based embryo model, progression from rosette formation to lumenogenesis accompanied by progression from naïve- to primed pluripotency was observed within Epi-like cells. Additionally, lineage specification of primordial germ cells and distal/anterior visceral endoderm-like cells was observed in epiblast- or visceral endoderm-like compartments, respectively. The system presented in this study allows for fast and reproducible generation of embryo-like structures, providing an additional tool to study aspects of early embryogenesis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. - PubMed
-
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. - PubMed
-
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
-
- Kunath T, et al. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–1661. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

