Deep dissection of the antiviral immune profile of patients with COVID-19
- PMID: 34916602
- PMCID: PMC8677724
- DOI: 10.1038/s42003-021-02852-1
Deep dissection of the antiviral immune profile of patients with COVID-19
Abstract
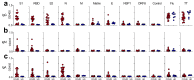
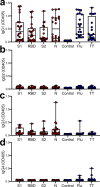
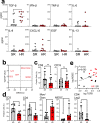
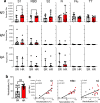
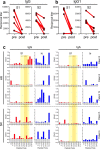
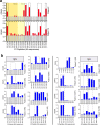
In light of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variants potentially undermining humoral immunity, it is important to understand the fine specificity of the antiviral antibodies. We screened 20 COVID-19 patients for antibodies against 9 different SARS-CoV-2 proteins observing responses against the spike (S) proteins, the receptor-binding domain (RBD), and the nucleocapsid (N) protein which were of the IgG1 and IgG3 subtypes. Importantly, mutations which typically occur in the B.1.351 "South African" variant, significantly reduced the binding of anti-RBD antibodies. Nine of 20 patients were critically ill and were considered high-risk (HR). These patients showed significantly higher levels of transforming growth factor beta (TGF-β) and myeloid-derived suppressor cells (MDSC), and lower levels of CD4+ T cells expressing LAG-3 compared to standard-risk (SR) patients. HR patients evidenced significantly higher anti-S1/RBD IgG antibody levels and an increased neutralizing activity. Importantly, a large proportion of S protein-specific antibodies were glycosylation-dependent and we identified a number of immunodominant linear epitopes within the S1 and N proteins. Findings derived from this study will not only help us to identify the most relevant component of the anti-SARS-CoV-2 humoral immune response but will also enable us to design more meaningful immunomonitoring methods for anti-COVID-19 vaccines.
© 2021. The Author(s).
Conflict of interest statement
C.R. was a speaker for Merck Sharp and Dohme, AstraZeneca, and Roche (CH) and has research collaborations (non-financial support) with Guardant Health; advisory board activity: Archer, Inivata and MD Serono, Novartis, and BMS. Research grant from LCRF-Pfizer. The rest of the authors declare no competing interests.
Figures

References
-
- World Health Organization. Coronavirus disease (COVID-19) pandemic. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

