Engineered HaloTag variants for fluorescence lifetime multiplexing
- PMID: 34916672
- PMCID: PMC8748199
- DOI: 10.1038/s41592-021-01341-x
Engineered HaloTag variants for fluorescence lifetime multiplexing
Abstract
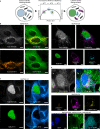
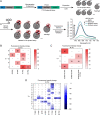
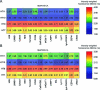
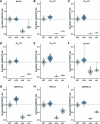
Self-labeling protein tags such as HaloTag are powerful tools that can label fusion proteins with synthetic fluorophores for use in fluorescence microscopy. Here we introduce HaloTag variants with either increased or decreased brightness and fluorescence lifetime compared with HaloTag7 when labeled with rhodamines. Combining these HaloTag variants enabled live-cell fluorescence lifetime multiplexing of three cellular targets in one spectral channel using a single fluorophore and the generation of a fluorescence lifetime-based biosensor. Additionally, the brightest HaloTag variant showed up to 40% higher brightness in live-cell imaging applications.
© 2021. The Author(s).
Conflict of interest statement
K.J. is inventor on patents filed by MPG and EPFL on fluorophores and labeling technologies. M.J.R. is an employee of Leica Microsystems. The remaining authors declare no competing interests.
Figures



References
-
- Gregor, I & Patting, M. Pattern-based linear unmixing for efficient and reliable analysis of multicomponent TCSPC data. In Advanced Photon Counting (eds Kapusta, P. et al.). 51–70. Springer International, 2013.
-
- Vallmitjana A, et al. Resolution of 4 components in the same pixel in FLIM images using the phasor approach. Methods Appl. Fluoresc. 2020;8:035001. - PubMed
-
- Niehörster T, et al. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat. Methods. 2016;13:257–262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

