Identification of the Potential Prognosis Biomarkers in Hepatocellular Carcinoma: An Analysis Based on WGCNA and PPI
- PMID: 34916837
- PMCID: PMC8670864
- DOI: 10.2147/IJGM.S338500
Identification of the Potential Prognosis Biomarkers in Hepatocellular Carcinoma: An Analysis Based on WGCNA and PPI
Abstract
Aim: This study was done to determine biomarkers for the prognostic prediction of hepatocellular carcinoma (HCC).
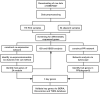
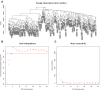
Materials and methods: In the Gene Expression Omnibus, the gene expression profiles of HCC were downloaded. Biomarkers were identified by weighted gene co-expression network analysis and protein-protein interaction network analysis.
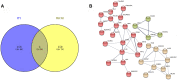
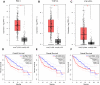
Results: There were 24 modules, which were characterized by the high correlation with HCC. Meanwhile, through enrichment analysis, differentially expressed genes were largely participated in the ubiquitination and autophagy processes. Moreover, PRC1, TOP2A and CKAP2L may be the hub genes involved in HCC tumorigenesis, and their biomarker roles were further demonstrated via Gene Expression Profiling Interactive Analysis (GEPIA) and Oncomine databases. In addition, the levels of PRC1, TOP2A and CKAP2L were obviously up-regulated in the sera of HCC patients.
Conclusion: PRC1, TOP2A and CKAP2L may serve as biomarkers for the prognostic prediction of HCC patients.
Keywords: PPI; WGCNA; biomarker; hepatocellular carcinoma; prognosis.
© 2021 Huang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

