Soybean Roots and Soil From High- and Low-Yielding Field Sites Have Different Microbiome Composition
- PMID: 34917042
- PMCID: PMC8669749
- DOI: 10.3389/fmicb.2021.675352
Soybean Roots and Soil From High- and Low-Yielding Field Sites Have Different Microbiome Composition
Abstract
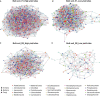
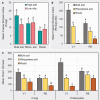
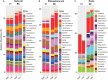
The occurrence of high- (H) and low- (L) yielding field sites within a farm is a commonly observed phenomenon in soybean cultivation. Site topography, soil physical and chemical attributes, and soil/root-associated microbial composition can contribute to this phenomenon. In order to better understand the microbial dynamics associated with each site type (H/L), we collected bulk soil (BS), rhizosphere soil (RS), and soybean root (R) samples from historically high and low yield sites across eight Pennsylvania farms at V1 (first trifoliate) and R8 (maturity) soybean growth stages (SGS). We extracted DNA extracted from collected samples and performed high-throughput sequencing of PCR amplicons from both the fungal ITS and prokaryotic 16S rRNA gene regions. Sequences were then grouped into amplicon sequence variants (ASVs) and subjected to network analysis. Based on both ITS and 16S rRNA gene data, a greater network size and edges were observed for all sample types from H-sites compared to L-sites at both SGS. Network analysis suggested that the number of potential microbial interactions/associations were greater in samples from H-sites compared to L-sites. Diversity analyses indicated that site-type was not a main driver of alpha and beta diversity in soybean-associated microbial communities. L-sites contained a greater percentage of fungal phytopathogens (ex: Fusarium, Macrophomina, Septoria), while H-sites contained a greater percentage of mycoparasitic (ex: Trichoderma) and entomopathogenic (ex: Metarhizium) fungal genera. Furthermore, roots from H-sites possessed a greater percentage of Bradyrhizobium and genera known to contain plant growth promoting bacteria (ex: Flavobacterium, Duganella). Overall, our results revealed that there were differences in microbial composition in soil and roots from H- and L-sites across a variety of soybean farms. Based on our findings, we hypothesize that differences in microbial composition could have a causative relationship with observed within-farm variability in soybean yield.
Keywords: bacterial 16S rRNA; fungal ITS; metabarcoding; metagenomics; microbiome; soybean; spatial yield variation.
Copyright © 2021 Bandara, Weerasooriya, Trexler, Bell and Esker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Aoki T., O’Donnell K., Scandiani M. M. (2005). Sudden death syndrome of soybean in South America is caused by four species of Fusarium: Fusarium brasiliense sp. nov., F. cuneirostrum sp. nov., F. tucumaniae, and F. virguliforme. Mycoscience 46 162–183. 10.1007/s10267-005-0235-y - DOI
-
- Bagwan N. B. (2010). Evaluation of Trichoderma compatibility with fungicides, pesticides, organic cakes and botanicals for integrated management of soil borne diseases of soybean [Glycine max (L.) Merril]. Int. J. Plant Prot. 3 206–209.
LinkOut - more resources
Full Text Sources
Research Materials

