m6A Regulator-Mediated Methylation Modification Patterns and Characteristics of Immunity in Blood Leukocytes of COVID-19 Patients
- PMID: 34917088
- PMCID: PMC8669770
- DOI: 10.3389/fimmu.2021.774776
m6A Regulator-Mediated Methylation Modification Patterns and Characteristics of Immunity in Blood Leukocytes of COVID-19 Patients
Abstract
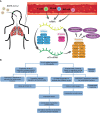
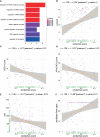
Both RNA N6-methyladenosine (m6A) modification of SARS-CoV-2 and immune characteristics of the human body have been reported to play an important role in COVID-19, but how the m6A methylation modification of leukocytes responds to the virus infection remains unknown. Based on the RNA-seq of 126 samples from the GEO database, we disclosed that there is a remarkably higher m6A modification level of blood leukocytes in patients with COVID-19 compared to patients without COVID-19, and this difference was related to CD4+ T cells. Two clusters were identified by unsupervised clustering, m6A cluster A characterized by T cell activation had a higher prognosis than m6A cluster B. Elevated metabolism level, blockage of the immune checkpoint, and lower level of m6A score were observed in m6A cluster B. A protective model was constructed based on nine selected genes and it exhibited an excellent predictive value in COVID-19. Further analysis revealed that the protective score was positively correlated to HFD45 and ventilator-free days, while negatively correlated to SOFA score, APACHE-II score, and crp. Our works systematically depicted a complicated correlation between m6A methylation modification and host lymphocytes in patients infected with SARS-CoV-2 and provided a well-performing model to predict the patients' outcomes.
Keywords: COVID-19; immune characteristics; leukocytes; m6A methylation modification; protective model.
Copyright © 2021 Qiu, Hua, Li, Zhou and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

