TranSNPs: A class of functional SNPs affecting mRNA translation potential revealed by fraction-based allelic imbalance
- PMID: 34917903
- PMCID: PMC8666669
- DOI: 10.1016/j.isci.2021.103531
TranSNPs: A class of functional SNPs affecting mRNA translation potential revealed by fraction-based allelic imbalance
Abstract
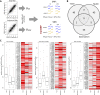
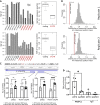
Few studies have explored the association between SNPs and alterations in mRNA translation potential. We developed an approach to identify SNPs that can mark allele-specific protein expression levels and could represent sources of inter-individual variation in disease risk. Using MCF7 cells under different treatments, we performed polysomal profiling followed by RNA sequencing of total or polysome-associated mRNA fractions and designed a computational approach to identify SNPs showing a significant change in the allelic balance between total and polysomal mRNA fractions. We identified 147 SNPs, 39 of which located in UTRs. Allele-specific differences at the translation level were confirmed in transfected MCF7 cells by reporter assays. Exploiting breast cancer data from TCGA we identified UTR SNPs demonstrating distinct prognosis features and altering binding sites of RNA-binding proteins. Our approach produced a catalog of tranSNPs, a class of functional SNPs associated with allele-specific translation and potentially endowed with prognostic value for disease risk.
Keywords: Computational bioinformatics; Molecular mechanism of gene regulation; Transcriptomics.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Andreotti V., Bisio A., Bressac-de Paillerets B., Harland M., Cabaret O., Newton-Bishop J., Pastorino L., Bruno W., Bertorelli R., De Sanctis V., et al. The CDKN2A/p16(INK) (4a) 5’UTR sequence and translational regulation: impact of novel variants predisposing to melanoma. Pigment Cell Melanoma Res. 2016;29:210–221. doi: 10.1111/pcmr.12444. - DOI - PubMed
-
- Andrysik Z., Galbraith M.D., Guarnieri A.L., Zaccara S., Sullivan K.D., Pandey A., MacBeth M., Inga A., Espinosa J.M. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res. 2017;27:1645–1657. doi: 10.1101/gr.220533.117. - DOI - PMC - PubMed
-
- Annunziato S., de Ruiter J.R., Henneman L., Brambillasca C.S., Lutz C., Vaillant F., Ferrante F., Drenth A.P., van der Burg E., Siteur B., et al. Comparative oncogenomics identifies combinations of driver genes and drug targets in BRCA1-mutated breast cancer. Nat. Commun. 2019;10:397. doi: 10.1038/s41467-019-08301-2. - DOI - PMC - PubMed
-
- Bailey S.D., Desai K., Kron K.J., Mazrooei P., Sinnott-Armstrong N.A., Treloar A.E., Dowar M., Thu K.L., Cescon D.W., Silvester J., et al. Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat. Genet. 2016;48:1260–1266. doi: 10.1038/ng.3650. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

