A high-throughput amplicon sequencing approach for population-wide species diversity and composition survey
- PMID: 34918473
- PMCID: PMC10234416
- DOI: 10.1111/1755-0998.13576
A high-throughput amplicon sequencing approach for population-wide species diversity and composition survey
Abstract
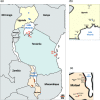
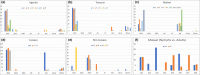
Management of agricultural pests requires an understanding of pest species diversity, their interactions with beneficial insects and spatial-temporal patterns of pest abundance. Invasive and agriculturally important insect pests can build up very high populations, especially in cropping landscapes. Traditionally, sampling effort for species identification involves small sample sizes and is labour intensive. Here, we describe a multiprimer high throughput sequencing (HTS) metabarcoding method and associated analytical workflow for a rapid, intensive, high-volume survey of pest species compositions. We demonstrate our method using the taxonomically challenging Bemisia pest cryptic species complex as examples. The whiteflies Bemisia including the"tabaci" species are agriculturally important capable of vectoring diverse plant viruses that cause diseases and crop losses. Our multiprimer metabarcoding HTS amplicon approach simultaneously process high volumes of whitefly individuals, with efficiency to detect rare (i.e., 1%) test-species, while our improved whitefly primers for metabarcoding also detected beneficial hymenopteran parasitoid species from whitefly nymphs. Field-testing our redesigned Bemisia metabarcoding primer sets across the Tanzania, Uganda and Malawi cassava cultivation landscapes, we identified the sub-Saharan Africa 1 Bemisia putative species as the dominant pest species, with other cryptic Bemisia species being detected at various abundances. We also provide evidence that Bemisia species compositions can be affected by host crops and sampling techniques that target either nymphs or adults. Our multiprimer HTS metabarcoding method incorporated two overlapping amplicons of 472 bp and 518 bp that spanned the entire 657 bp 3' barcoding region for Bemisia, and is particularly suitable to molecular diagnostic surveys of this highly cryptic insect pest species complex that also typically exhibited high population densities in heavy crop infestation episodes. Our approach can be adopted to understand species biodiversity across landscapes, with broad implications for improving transboundary biosecurity preparedness, thus contributing to molecular ecological knowledge and the development of control strategies for high-density, cryptic, pest-species complexes.
Keywords: Bemisia; African cassava whitefly; Aphelinidae; metabarcoding; multiprimer molecular diagnostics; parasitoids.
© 2021 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ally, H. M. , El Hamss, H. , Simiand, C. , Maruthi, M. N. , Colvin, J. , Omongo, C. A. , & Delatte, H. (2019). What has changed in the outbreaking populations of the severe crop pest whitefly species in cassava in two decades? Scientific Reports, 9, 14796. 10.1038/s41598-019-50259-0 - DOI - PMC - PubMed
-
- Arribas, P. , Andújar, C. , Bidartondo, M. I. , Bohmann, K. , Coissac, É. , Creer, S. , deWaard, J. R. , Elbrecht, V. , Ficetola, G. F. , Goberna, M. , Kennedy, S. , Krehenwinkel, H. , Leese, F. , Novotny, V. , Ronquist, F. , Yu, D. W. , Zinger, L. , Creedy, T. J. , Meramveliotakis, E. , … Emerson, B. C. (2021). Connecting high‐throughput biodiversity inventories: Opportunities for a site‐based genomic framework for global integration and synthesis. Molecular Ecology, 30(5), 1120–1135. 10.1111/mec.15797 - DOI - PMC - PubMed
-
- Bansch, S. , Tscharntke, T. , Wunschiers, R. , Netter, L. , Brenig, B. , Gabriel, D. , & Westphal, C. (2020). Using ITS2 metabarcoding and microscopy to analyse shifts in pollen diets of honey bees and bumble bees along a mass‐flowering crop gradient. Molecular Ecology, 29(24), 5003–5018. 10.1111/mec.15675 - DOI - PubMed
-
- Berry, S. D. , Fondong, V. N. , Rey, C. , Rogan, D. , Fauquet, C. M. , & Brown, J. K. (2004). Molecular Evidence for Five Distinct Bemisia tabaci (Homoptera: Aleyrodidae) Geographic Haplotypes Associated with Cassava Plants in Sub‐Saharan Africa. Annals of the Entomological Society of America, 97(4), 852–859. 10.1603/0013-8746(2004)097[0852:meffdb]2.0.co;2 - DOI
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

