exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids
- PMID: 34918744
- PMCID: PMC8728150
- DOI: 10.1093/nar/gkab1085
exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids
Abstract
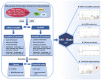
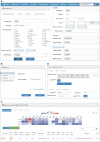
Extracellular vesicles (EVs) are small membranous vesicles that contain an abundant cargo of different RNA species with specialized functions and clinical implications. Here, we introduce an updated online database (http://www.exoRBase.org), exoRBase 2.0, which is a repository of EV long RNAs (termed exLRs) derived from RNA-seq data analyses of diverse human body fluids. In exoRBase 2.0, the number of exLRs has increased to 19 643 messenger RNAs (mRNAs), 15 645 long non-coding RNAs (lncRNAs) and 79 084 circular RNAs (circRNAs) obtained from ∼1000 human blood, urine, cerebrospinal fluid (CSF) and bile samples. Importantly, exoRBase 2.0 not only integrates and compares exLR expression profiles but also visualizes the pathway-level functional changes and the heterogeneity of origins of circulating EVs in the context of different physiological and pathological conditions. Our database provides an attractive platform for the identification of novel exLR signatures from human biofluids that will aid in the discovery of new circulating biomarkers to improve disease diagnosis and therapy.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes.Nucleic Acids Res. 2018 Jan 4;46(D1):D106-D112. doi: 10.1093/nar/gkx891. Nucleic Acids Res. 2018. PMID: 30053265 Free PMC article.
-
Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis.Clin Chem. 2019 Jun;65(6):798-808. doi: 10.1373/clinchem.2018.301291. Epub 2019 Mar 26. Clin Chem. 2019. PMID: 30914410
-
Extracellular vesicles long RNA profiling identifies abundant mRNA, circRNA and lncRNA in human bile as potential biomarkers for cancer diagnosis.Carcinogenesis. 2023 Dec 2;44(8-9):671-681. doi: 10.1093/carcin/bgad063. Carcinogenesis. 2023. PMID: 37696683
-
Clinical applications of extracellular vesicle long RNAs.Crit Rev Clin Lab Sci. 2020 Dec;57(8):508-521. doi: 10.1080/10408363.2020.1751584. Epub 2020 May 12. Crit Rev Clin Lab Sci. 2020. PMID: 32393091 Review.
-
Circulating RNAs in prostate cancer patients.Cancer Lett. 2022 Jan 1;524:57-69. doi: 10.1016/j.canlet.2021.10.011. Epub 2021 Oct 14. Cancer Lett. 2022. PMID: 34656688 Review.
Cited by
-
Exploring Circular RNA Profile and Expression in Extracellular Vesicles.Methods Mol Biol. 2024;2765:47-59. doi: 10.1007/978-1-0716-3678-7_3. Methods Mol Biol. 2024. PMID: 38381333
-
Benchmarking transcriptome deconvolution methods for estimating tissue- and cell-type-specific extracellular vesicle abundances.J Extracell Vesicles. 2024 Sep;13(9):e12511. doi: 10.1002/jev2.12511. J Extracell Vesicles. 2024. PMID: 39320021 Free PMC article.
-
Genetic prediction of blood metabolites mediating the relationship between gut microbiota and Alzheimer's disease: a Mendelian randomization study.Front Microbiol. 2024 Aug 19;15:1414977. doi: 10.3389/fmicb.2024.1414977. eCollection 2024. Front Microbiol. 2024. PMID: 39224217 Free PMC article.
-
Identifying diagnostic markers and constructing a prognostic model for small-cell lung cancer based on blood exosome-related genes and machine-learning methods.Front Oncol. 2022 Dec 22;12:1077118. doi: 10.3389/fonc.2022.1077118. eCollection 2022. Front Oncol. 2022. PMID: 36620585 Free PMC article.
-
Host Long Noncoding RNAs as Key Players in Mycobacteria-Host Interactions.Microorganisms. 2024 Dec 21;12(12):2656. doi: 10.3390/microorganisms12122656. Microorganisms. 2024. PMID: 39770858 Free PMC article. Review.
References
-
- Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K.et al. .. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018; 7:1535750. - PMC - PubMed
-
- Bordanaba-Florit G., Royo F., Kruglik S.G., Falcon-Perez J.M.. Using single-vesicle technologies to unravel the heterogeneity of extracellular vesicles. Nat. Protoc. 2021; 16:3163–3185. - PubMed
-
- Murillo O.D., Thistlethwaite W., Rozowsky J., Subramanian S.L., Lucero R., Shah N., Jackson A.R., Srinivasan S., Chung A., Laurent C.D.et al. .. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019; 177:463–477. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

