Whole genome sequencing-based copy number variations reveal novel pathways and targets in Alzheimer's disease
- PMID: 34918867
- PMCID: PMC9264340
- DOI: 10.1002/alz.12507
Whole genome sequencing-based copy number variations reveal novel pathways and targets in Alzheimer's disease
Abstract
Introduction: A few copy number variations (CNVs) have been reported for Alzheimer's disease (AD). However, there is a lack of a systematic investigation of CNVs in AD based on whole genome sequencing (WGS) data.
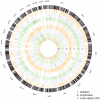
Methods: We used four methods to identify consensus CNVs from the WGS data of 1,411 individuals and further investigated their functional roles in AD using the matched transcriptomic and clinicopathological data.
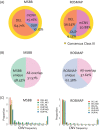
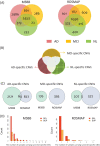
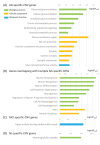
Results: We identified 3,012 rare AD-specific CNVs whose residing genes are enriched for cellular glucuronidation and neuron projection pathways. Genes whose mRNA expressions are significantly correlated with common CNVs are involved in major histocompatibility complex class II receptor activity. Integration of CNVs, gene expression, and clinical and pathological traits further pinpoints a key CNV that potentially regulates immune response in AD.
Discussion: We identify CNVs as potential genetic regulators of immune response in AD. The identified CNVs and their downstream gene networks reveal novel pathways and targets for AD.
Keywords: Alzheimer's disease; copy number variation; correlation network; immune response; late-onset Alzheimer's disease; multi-omics integration; regulation of response to external stimulus; whole genomic sequencing.
© 2021 The Authors. Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Chen Ming, Minghui Wang, Qian Wang, Ryan Neff, Erming Wang, Qi Shen, Joseph S. Reddy, and Xue Wang have nothing to disclose. Bin Zhang, Mariet Allen, David A. Bennett, Vahram Haroutunian, and Eric Schadt received support for the present manuscript. Specifically, this manuscript was supported by Bin Zhang‘s NIH/NIA grants (R01AG046170, RF1AG054014, RF1AG057440, R01AG057907, U01AG052411, R01AG062355, U01AG058635, R01AG068030) and Mariet Allen's grant (U01‐AG046139) as well as NIH grants for David A. Bennett, Vahram Haroutunian, and Eric Schadt. In the past 36 months, Bin Zhang, Minghui Wang, Mariet Allen, Nilüfer Ertekin‐Taner, Philip L. De Jager, David A. Bennett, Vahram Haroutunian, and Eric Schadt received grants from NIH or foundations. Specifically, Bin Zhang received 21 grants (R01AG046170, RF1AG054014, RF1AG057440, R01AG057907, U01AG052411, R01AG062355, U01AG058635, R01AG068030, R01AG062661, R01AG062355, HHS‐NIH‐NIAID‐BAA2018, R01 MH111679, R01DA043247, R01DK118243, R01DA048279, R56AG058655, R01AG063819, R01D029322, R01DA047880, R01AG062661, R01AG060341, R21AI149013). Mariet Allen received 3 grants (U01‐AG046139, R01‐AG061796, U01‐AG046139, RF1‐AG51504). Nilüfer Ertekin‐Taner received 7 grants (U01AG046139, RF1AG051504, R01AG061796, P30AG062677, U01AG061359, NHLBI75N92019D00031/75N92019F00125, R01AG050603). Minghui Wang received one grant (1RF1AG066526‐01A1). Philip L. De Jager, David A. Bennett, Vahram Haroutunian, and Eric Schadt received NIH grants. David A. Bennett received a grant from NIH Neurovision. Eric Schadt also received grants from the Helmsley Foundation. None of the co‐authors received royalties or licenses in the past 36 months. In the past 36 months, Philip L. De Jager received consultant fees from Partech, Roche, and Biogen; David A. Bennett received consultant fees from AbbVie, DSMB, Takeda, Origent, and SBIR; Vahram Haroutunian received a consultant fee of $350 from Synaptec; and Eric Schadt is paid by Berg Pharmaceuticals for participation on their scientific advisory board. In the past 36 months, Bin Zhang received an honorarium for presentation from Lehigh University; Nilüfer Ertekin‐Taner received payments for her presentations at the 15th International Symposium on Geriatrics and Gerontology Inflammation and Dementia: Genomics, System and Therapeutics, Nagoya, Japan; Philip L. De Jager received payments from Novartis, Astra Zeneca, and Biogen for lectures/presentations; David A. Bennett received payments or honoraria from academia in US and government (NGO) for lectures/presentations. In the past 36 months, Bin Zhang received support from FNIH to attend the AMP‐AD program meeting at NIH (the payment was made to him); Minghui Wang received support from FNIH for his travel and hotel lodging for attending NIH project meetings (the payments were made to him); Mariet Allen received a travel fellowship from Alzheimer's Association International Conference for conference registration but without payments, travel reimbursement from Alzheimer's Association, and travel reimbursement from NIH (the payments were made to her); Nilüfer Ertekin‐Taner received supports for attending the following conferences: 11th ISABS Conference (Split, Croatia), the 15th International Symposium on Geriatrics and Gerontology Inflammation and Dementia: Genomics, System and Therapeutics (Nagoya, Japan), multiple NIH meetings, Department of Neurology, Indiana University School of Medicine (Bloomington, Indiana); David A. Bennett received support for attending meetings from academia in US and government (NGO). In the past 36 months, the following authors have pending patents: Bin Zhang (US2020/056080 titled “STATHMIN 2 (STMN2) as a therapeutic target for Parkinson's disease,” MS‐0029‐01‐US‐P titled “A Novel therapeutic strategy for targeting the molecular subtypes of AD,” and MS‐00‐30‐01‐US‐P titled “Novel Compounds for Treating Alzheimer's Disease”), Minghui Wang (US2020/056080 titled “STATHMIN 2 (STMN2) as a therapeutic target for Parkinson's disease”), Ryan Neff (MS‐0029‐01‐US‐P titled “A Novel therapeutic strategy for targeting the molecular subtypes of AD”). Eric Schadt has patents under consideration relating to novel drug targets and targeting mechanisms for AD, patent applications filed and under review for biomarkers relating to various forms of cancer, predicting drug response and matching to therapies. In the past 36 months, Eric Schadt was on the board of directors for Sema4, a for‐profit company, and for Sage Bionetworks and 4YouAndMe, both non‐profit research institutions; Nilüfer Ertekin‐Taner was on the external advisory board for the NIH TREAT‐AD consortium; David A. Bennett was on the external advisory board of AbbVie.
David A. Bennett received equipment, materials, drugs, medical writing, gifts, or other services from Rush philanthropy in the past 36 months. The authors declare that they have no other competing interests.
Figures




References
-
- Patterson C. World Alzheimer Report 2018: The State of The Art of Dementia Research: New Frontiers. London, UK: Alzheimer's Disease International (ADI); 2018.
-
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer's disease. Arch Gen Psychiat. 2006;63(2):168‐174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AG072975/AG/NIA NIH HHS/United States
- RF1AG054014/AG/NIA NIH HHS/United States
- U01AG46152/AG/NIA NIH HHS/United States
- R01AG15819/AG/NIA NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- RF1 AG059319/AG/NIA NIH HHS/United States
- R01 AG068030/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- R01 AG061796/AG/NIA NIH HHS/United States
- RF1AG057440/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- P30 AG066514/AG/NIA NIH HHS/United States
- U01AG052411/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- U01AG058635/AG/NIA NIH HHS/United States
- RF1 AG057440/AG/NIA NIH HHS/United States
- P30AG10161/AG/NIA NIH HHS/United States
- RF1 AG054014/AG/NIA NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- U01 AG052411/AG/NIA NIH HHS/United States
- U01AG046170/AG/NIA NIH HHS/United States
- R01 AG057907/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- R01AG057907/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- R01AG068030/AG/NIA NIH HHS/United States
- R01AG062355/AG/NIA NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- R01AG17917/AG/NIA NIH HHS/United States
- U01AG61356/AG/NIA NIH HHS/United States
- U01 AG058635/AG/NIA NIH HHS/United States
- R01 AG062355/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

