Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer
- PMID: 34919143
- PMCID: PMC8932635
- DOI: 10.1084/jem.20210564
Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer
Abstract
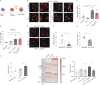
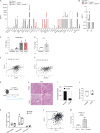
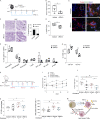
Tumor-associated macrophages (TAMs) are correlated with the progression of prostatic adenocarcinoma (PCa). The mechanistic basis of this correlation and therapeutic strategies to target TAMs in PCa remain poorly defined. Here, single-cell RNA sequencing was used to profile the transcriptional landscape of TAMs in human PCa, leading to identification of a subset of macrophages characterized by dysregulation in transcriptional pathways associated with lipid metabolism. This subset of TAMs correlates positively with PCa progression and shorter disease-free survival and is characterized by an accumulation of lipids that is dependent on Marco. Mechanistically, cancer cell-derived IL-1β enhances Marco expression on macrophages, and reciprocally, cancer cell migration is promoted by CCL6 released by lipid-loaded TAMs. Moreover, administration of a high-fat diet to tumor-bearing mice raises the abundance of lipid-loaded TAMs. Finally, targeting lipid accumulation by Marco blockade hinders tumor growth and invasiveness and improves the efficacy of chemotherapy in models of PCa, pointing to combinatorial strategies that may influence patient outcomes.
© 2021 Masetti et al.
Conflict of interest statement
Disclosures: R.A. DePinho reported being a Founder and Advisor for Tvardi Therapeutics, Asylia Therapeutics, Nirogy Therapeutics, Stellanova Therapeutics, and Sporos Bioventures. The focus of these companies is not directly related to the content of this manuscript. S. Gordon reported personal fees from Verseau, Myeloid Therapeutics, and Alnylam outside the submitted work. No other disclosures were reported.
Figures






References
-
- Alimonti, A., Nardella C., Chen Z., Clohessy J.G., Carracedo A., Trotman L.C., Cheng K., Varmeh S., Kozma S.C., Thomas G., et al. . 2010. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J. Clin. Invest. 120:681–693. 10.1172/JCI40535 - DOI - PMC - PubMed
-
- Balaban, S., Nassar Z.D., Zhang A.Y., Hosseini-Beheshti E., Centenera M.M., Schreuder M., Lin H.M., Aishah A., Varney B., Liu-Fu F., et al. . 2019. Extracellular fatty acids are the major contributor to lipid synthesis in prostate cancer. Mol. Cancer Res. 17:949–962. 10.1158/1541-7786.MCR-18-0347 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

