Flooding and ecological restoration promote wetland microbial communities and soil functions on former cranberry farmland
- PMID: 34919560
- PMCID: PMC8683025
- DOI: 10.1371/journal.pone.0260933
Flooding and ecological restoration promote wetland microbial communities and soil functions on former cranberry farmland
Abstract
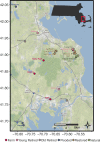
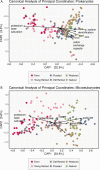
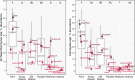
Microbial communities are early responders to wetland degradation, and instrumental players in the reversal of this degradation. However, our understanding of soil microbial community structure and function throughout wetland development remains incomplete. We conducted a survey across cranberry farms, young retired farms, old retired farms, flooded former farms, ecologically restored former farms, and natural reference wetlands with no history of cranberry farming. We investigated the relationship between the microbial community and soil characteristics that restoration intends to maximize, such as soil organic matter, cation exchange capacity and denitrification potential. Among the five treatments considered, flooded and restored sites had the highest prokaryote and microeukaryote community similarity to natural wetlands. In contrast, young retired sites had similar communities to farms, and old retired sites failed to develop wetland microbial communities or functions. Canonical analysis of principal coordinates revealed that soil variables, in particular potassium base saturation, sodium, and denitrification potential, explained 45% of the variation in prokaryote communities and 44% of the variation in microeukaryote communities, segregating soil samples into two clouds in ordination space: farm, old retired and young retired sites on one side and restored, flooded, and natural sites on the other. Heat trees revealed possible prokaryotic (Gemmatimonadetes) and microeukaryotic (Rhizaria) indicators of wetland development, along with a drop in the dominance of Nucletmycea in restored sites, a class that includes suspected mycorrhizal symbionts of the cranberry crop. Flooded sites showed the strongest evidence of wetland development, with triple the soil organic matter accumulation, double the cation exchange capacity, and seventy times the denitrification potential compared to farms. However, given that flooding does not promote any of the watershed or habitat benefits as ecological restoration, we suggest that flooding can be used to stimulate beneficial microbial communities and soil functions during the restoration waiting period, or when restoration is not an option.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Strickland MS, Callaham MA, Gardiner ES, Stanturf JA, Leff JW, et al.. Response of soil microbial community composition and function to a bottomland forest restoration intensity gradient. Appl Soil Ecol. 2017;119:317–326.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

