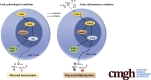
TOB1 Blocks Intestinal Mucosal Inflammation Through Inducing ID2-Mediated Suppression of Th1/Th17 Cell Immune Responses in IBD
- PMID: 34920145
- PMCID: PMC8881672
- DOI: 10.1016/j.jcmgh.2021.12.007
TOB1 Blocks Intestinal Mucosal Inflammation Through Inducing ID2-Mediated Suppression of Th1/Th17 Cell Immune Responses in IBD
Abstract
Background & aims: TOB1 is an anti-proliferative protein of Tob/BTG family and typically involved in the tumorigenesis and T cell activation. Although TOB1 is associated with T helper 17 cell-related autoimmunity, its role in modulating T cell-mediated immune responses in IBD remains poorly understood. Here, we explored its expression and the underlying mechanisms involved in the pathogenesis of inflammatory bowel disease (IBD).
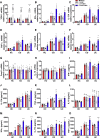
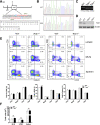
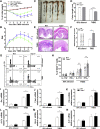
Methods: TOB1 and ID2 expression in IBD patients was examined by quantitative real time polymerase chain reaction and immunohistochemistry. IBD CD4+ T cells were transfected with lentivirus expressing TOB1, ID2, TOB1 short hairpin RNA and ID2 short hairpin RNA, respectively, and Tob1-/-CD4+ T cells were transfected with lentivirus expressing Id2. Experimental colitis was established in Tob1-/- mice by trinitrobenzene sulfonic acid enema and in Rag1-/- mice reconstituted with Tob1-/-CD45RBhighCD4+ T cells to further explore the role of Tob1 in intestinal mucosal inflammation. Splenic CD4+ T cells of Tob1-/- mice were sorted to determine transcriptome differences by RNA sequencing.
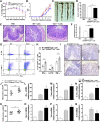
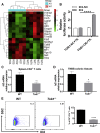
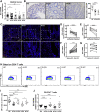
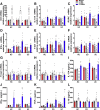
Results: TOB1 expression was decreased in inflamed mucosa and peripheral blood CD4+ T cells of IBD patients compared with healthy subjects. Overexpression of TOB1 downregulated IBD CD4+ T cells to differentiate into Th1/Th17 cells compared with control subjects. Severe colitis was observed in Tob1-/- mice through trinitrobenzene sulfonic acid enema or in Rag1-/- mice reconstituted with Tob1-/-CD45RBhighCD4+ T cells, compared with control animals. RNA sequencing analysis revealed ID2 as functional target of TOB1 to inhibit IBD CD4+ T cell differentiation into Th1/Th17 cells. Mechanistically, TOB1 was associated with Smad4/5 to induce ID2 expression and restrain Th1/Th17 cell differentiation.
Conclusions: TOB1 restrains intestinal mucosal inflammation through suppressing Th1/Th17 cell-mediated immune responses via the Smad4/5-ID2 pathway. It may serve as a novel therapeutic target for treatment of human IBD.
Keywords: CD4(+) T Cells; ID2; Inflammatory Bowel Disease; Mucosal Inflammation; TOB1.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Chang J.T. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383:2652–2664. - PubMed
-
- Maloy K.J., Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. - PubMed
-
- Park J.H., Peyrin-Biroulet L., Eisenhut M., Shin J.I. IBD immunopathogenesis: A comprehensive review of inflammatory molecules. Autoimmun Rev. 2017;16:416–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

