A Randomized Phase II Study Comparing Nivolumab with Carboplatin-Pemetrexed for EGFR-Mutated NSCLC with Resistance to EGFR Tyrosine Kinase Inhibitors (WJOG8515L)
- PMID: 34921023
- PMCID: PMC9397372
- DOI: 10.1158/1078-0432.CCR-21-3194
A Randomized Phase II Study Comparing Nivolumab with Carboplatin-Pemetrexed for EGFR-Mutated NSCLC with Resistance to EGFR Tyrosine Kinase Inhibitors (WJOG8515L)
Abstract
Purpose: Although the efficacy of programmed cell death-1 (PD-1) blockade is generally poor for non-small cell lung cancer (NSCLC) with activating mutations of the epidermal growth factor receptor (EGFR) gene, EGFR tyrosine kinase inhibitors (TKIs) may improve the tumor immune microenvironment. We performed a randomized study to assess whether nivolumab improves outcome compared with chemotherapy in such patients previously treated with EGFR-TKIs.
Patients and methods: Patients with EGFR-mutated NSCLC who acquired EGFR-TKI resistance not due to a secondary T790M mutation of EGFR were randomized 1:1 to nivolumab (n = 52) or carboplatin-pemetrexed (n = 50). The primary endpoint was progression-free survival (PFS).
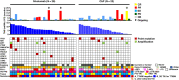
Results: Median PFS and 1-year PFS probability were 1.7 months and 9.6% for nivolumab versus 5.6 months and 14.0% for carboplatin-pemetrexed [log-rank P < 001; hazard ratio (HR) of 1.92, with a 60% confidence interval (CI) of 1.61-2.29]. Overall survival was 20.7 and 19.9 months [HR, 0.88 (95% CI, 0.53-1.47)], and response rate was 9.6% and 36.0% for nivolumab and carboplatin-pemetrexed, respectively. No subgroup including patients with a high tumor mutation burden showed a substantially longer PFS with nivolumab than with carboplatin-pemetrexed. The T-cell-inflamed gene expression profile score (0.11 vs. -0.17, P = 0.036) and expression of genes related to cytotoxic T lymphocytes or their recruitment were higher in tumors that showed a benefit from nivolumab.
Conclusions: Nivolumab did not confer a longer PFS compared with carboplatin-pemetrexed in the study patients. Gene expression profiling identified some cases with a favorable tumor immune microenvironment that was associated with nivolumab efficacy.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- World Health Organization. Cancer - Key Facts. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer; 2021.
-
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. . Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121–8. - PubMed
-
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. . Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

