Microbiome-mediated incapacitation of interferon lambda production in the oral mucosa
- PMID: 34921113
- PMCID: PMC8713781
- DOI: 10.1073/pnas.2105170118
Microbiome-mediated incapacitation of interferon lambda production in the oral mucosa
Erratum in
-
Correction for Rodriguez-Hernandez et al., Microbiome-mediated incapacitation of interferon lambda production in the oral mucosa.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2212111119. doi: 10.1073/pnas.2212111119. Epub 2022 Aug 17. Proc Natl Acad Sci U S A. 2022. PMID: 35976805 Free PMC article. No abstract available.
Abstract
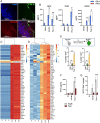
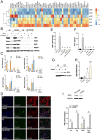
Here, we show that Porphyromonas gingivalis (Pg), an endogenous oral pathogen, dampens all aspects of interferon (IFN) signaling in a manner that is strikingly similar to IFN suppression employed by multiple viral pathogens. Pg suppressed IFN production by down-regulating several IFN regulatory factors (IRFs 1, 3, 7, and 9), proteolytically degrading STAT1 and suppressing the nuclear translocation of the ISGF3 complex, resulting in profound and systemic repression of multiple interferon-stimulated genes. Pg-induced IFN paralysis was not limited to murine models but was also observed in the oral tissues of human periodontal disease patients, where overabundance of Pg correlated with suppressed IFN generation. Mechanistically, multiple virulence factors and secreted proteases produced by Pg transcriptionally suppressed IFN promoters and also cleaved IFN receptors, making cells refractory to exogenous IFN and inducing a state of broad IFN paralysis. Thus, our data show a bacterial pathogen with equivalence to viruses in the down-regulation of host IFN signaling.
Keywords: Interferon lambda; Porphyromonas gingivalis; oral epithelial cells; periodontitis; viral infection.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Puletic M., Popovic B., Jankovic S., Brajovic G., Detection rates of periodontal bacteria and herpesviruses in different forms of periodontal disease. Microbiol. Immunol. 64, 815–824 (2020). - PubMed
-
- Christersson L. A., Fransson C. L., Dunford R. G., Zambon J. J., Subgingival distribution of periodontal pathogenic microorganisms in adult periodontitis. J. Periodontol. 63, 418–425 (1992). - PubMed
-
- Imbronito A. V., Okuda O. S., Maria de Freitas N., Moreira Lotufo R. F., Nunes F. D., Detection of herpesviruses and periodontal pathogens in subgingival plaque of patients with chronic periodontitis, generalized aggressive periodontitis, or gingivitis. J. Periodontol. 79, 2313–2321 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

