Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing
- PMID: 34921143
- PMCID: PMC8683505
- DOI: 10.1038/s41467-021-27619-4
Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing
Abstract
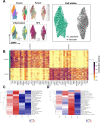
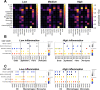
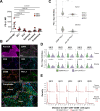
Head and neck squamous cell carcinoma (HNSCC) is characterized by complex relations between stromal, epithelial, and immune cells within the tumor microenvironment (TME). To enable the development of more efficacious therapies, we aim to study the heterogeneity, signatures of unique cell populations, and cell-cell interactions of non-immune and immune cell populations in 6 human papillomavirus (HPV)+ and 12 HPV- HNSCC patient tumor and matched peripheral blood specimens using single-cell RNA sequencing. Using this dataset of 134,606 cells, we show cell type-specific signatures associated with inflammation and HPV status, describe the negative prognostic value of fibroblasts with elastic differentiation specifically in the HPV+ TME, predict therapeutically targetable checkpoint receptor-ligand interactions, and show that tumor-associated macrophages are dominant contributors of PD-L1 and other immune checkpoint ligands in the TME. We present a comprehensive single-cell view of cell-intrinsic mechanisms and cell-cell communication shaping the HNSCC microenvironment.
© 2021. The Author(s).
Conflict of interest statement
R.L.F: Aduro Biotech, Inc; EMD Serono; MacroGenics, Inc.; Numab Therapeutics AG; Pfizer; Sanofi Tesaro; Zymeworks, Inc (honoraria); Astra-Zeneca/MedImmune (clinical trial, research funding); Bristol-Myers Squibb (honoraria, clinical trial, research funding); Merck (honoraria, clinical trial); Novasenta (honoraria, stock, research funding); (research funding). D.A.A.V.: Stock: Novasenta, Tizona, Trishula, Oncorus, Werewolf, Apeximmune; Consultancy: Tizona, Werewolf, F-Star, Astellas, BMS, MPM, Incyte, Bicara, Apeximmune, G1 Therapeutics, Innovent Bio, Kronos Bio; Grants: BMS, Astellas, Novasenta; Patents licensed and Royalties: Astellas, BMS, Novasenta. The remaining authors declare no competing interests.
Figures

References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Iacobuzio-Donahue CA, Litchfield K, Swanton C. Intratumor heterogeneity reflects clinical disease course. Nat. Cancer. 2020;1:3–6. - PubMed
-
- Chow LQM. Head and neck cancer. N. Engl. J. Med. 2020;382:60–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

