A targetable LIFR-NF-κB-LCN2 axis controls liver tumorigenesis and vulnerability to ferroptosis
- PMID: 34921145
- PMCID: PMC8683481
- DOI: 10.1038/s41467-021-27452-9
A targetable LIFR-NF-κB-LCN2 axis controls liver tumorigenesis and vulnerability to ferroptosis
Abstract
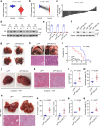
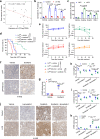
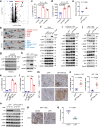
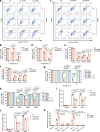
The growing knowledge of ferroptosis has suggested the role and therapeutic potential of ferroptosis in cancer, but has not been translated into effective therapy. Liver cancer, primarily hepatocellular carcinoma (HCC), is highly lethal with limited treatment options. LIFR is frequently downregulated in HCC. Here, by studying hepatocyte-specific and inducible Lifr-knockout mice, we show that loss of Lifr promotes liver tumorigenesis and confers resistance to drug-induced ferroptosis. Mechanistically, loss of LIFR activates NF-κB signaling through SHP1, leading to upregulation of the iron-sequestering cytokine LCN2, which depletes iron and renders insensitivity to ferroptosis inducers. Notably, an LCN2-neutralizing antibody enhances the ferroptosis-inducing and anticancer effects of sorafenib on HCC patient-derived xenograft tumors with low LIFR expression and high LCN2 expression. Thus, anti-LCN2 therapy is a promising way to improve liver cancer treatment by targeting ferroptosis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Villanueva A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019;380:1450–1462. - PubMed
-
- Roayaie S, et al. The role of hepatic resection in the treatment of hepatocellular cancer. Hepatology. 2015;62:440–451. - PubMed
-
- Mazzaferro V, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 1996;334:693–699. - PubMed
-
- Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol. 2016;65:1031–1042. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

