Co-option of an extracellular protease for transcriptional control of nutrient degradation in the fungus Aspergillus nidulans
- PMID: 34921231
- PMCID: PMC8683493
- DOI: 10.1038/s42003-021-02925-1
Co-option of an extracellular protease for transcriptional control of nutrient degradation in the fungus Aspergillus nidulans
Abstract
Nutrient acquisition is essential for all organisms. Fungi regulate their metabolism according to environmental nutrient availability through elaborate transcription regulatory programs. In filamentous fungi, a highly conserved GATA transcription factor AreA and its co-repressor NmrA govern expression of genes involved in extracellular breakdown, uptake, and metabolism of nitrogen nutrients. Here, we show that the Aspergillus nidulans PnmB protease is a moonlighting protein with extracellular and intracellular functions for nitrogen acquisition and metabolism. PnmB serves not only as a secreted protease to degrade extracellular nutrients, but also as an intracellular protease to control the turnover of the co-repressor NmrA, accelerating AreA transcriptional activation upon nitrogen starvation. PnmB expression is controlled by AreA, which activates a positive feedback regulatory loop. Hence, we uncover a regulatory mechanism in the well-established controls determining the response to nitrogen starvation, revealing functional evolution of a protease gene for transcriptional regulation and extracellular nutrient breakdown.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
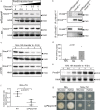

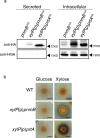

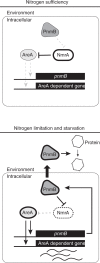
References
-
- St Leger RJ, Joshi L, Roberts DW. Adaptation of proteases and carbohydrases of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology. 1997;143:1983–1992. - PubMed
-
- Hu G, St Leger RS. A phylogenomic approach to reconstructing the diversification of serine proteases in fungi. J. Evol. Biol. 2004;17:1204–1214. - PubMed
-
- Katz ME, Buckland R, Hunter CC, Todd RB. Distinct roles for the p53-like transcription factor XprG and autophagy genes in the response to starvation. Fungal Genet. Biol. 2015;83:10–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

