Mahogunin Ring Finger 1 regulates pigmentation by controlling the pH of melanosomes in melanocytes and melanoma cells
- PMID: 34921635
- PMCID: PMC8738503
- DOI: 10.1007/s00018-021-04053-9
Mahogunin Ring Finger 1 regulates pigmentation by controlling the pH of melanosomes in melanocytes and melanoma cells
Abstract
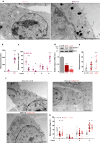
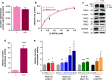
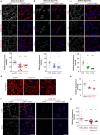
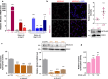
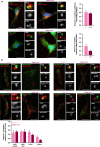
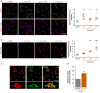
Mahogunin Ring Finger 1 (MGRN1) is an E3-ubiquitin ligase absent in dark-furred mahoganoid mice. We investigated the mechanisms of hyperpigmentation in Mgrn1-null melan-md1 melanocytes, Mgrn1-KO cells obtained by CRISPR-Cas9-mediated knockdown of Mgrn1 in melan-a6 melanocytes, and melan-a6 cells depleted of MGRN1 by siRNA treatment. Mgrn1-deficient melanocytes showed higher melanin content associated with increased melanosome abundance and higher fraction of melanosomes in highly melanized maturation stages III-IV. Expression, post-translational processing and enzymatic activity of the rate-limiting melanogenic enzyme tyrosinase measured in cell-free extracts were comparable in control and MGRN1-depleted cells. However, tyrosinase activity measured in situ in live cells and expression of genes associated with regulation of pH increased upon MGRN1 repression. Using pH-sensitive fluorescent probes, we found that downregulation of MGRN1 expression in melanocytes and melanoma cells increased the pH of acidic organelles, including melanosomes, strongly suggesting a previously unknown role of MGRN1 in the regulation of melanosomal pH. Among the pH regulatory genes upregulated by Mgrn1 knockdown, we identified those encoding several subunits of the vacuolar adenosine triphosphatase V-ATPase (mostly Atp6v0d2) and a calcium channel of the transient receptor potential channel family, Mucolipin 3 (Mcoln3). Manipulation of expression of the Mcoln3 gene showed that overexpression of Mcoln3 played a significant role in neutralization of the pH of acidic organelles and activation of tyrosinase in MGRN1-depleted cells. Therefore, lack of MGRN1 led to cell-autonomous stimulation of pigment production in melanocytes mostly by increasing tyrosinase specific activity through neutralization of the melanosomal pH in a MCOLN3-dependent manner.
Keywords: Lysosome-related organelles; Mahogunin Ring Finger 1 (MGRN1); Melanin; Melanosomal pH; Mucolipin 3 (MCOLN3); Tyrosinase.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Melanosomal pH controls rate of melanogenesis, eumelanin/phaeomelanin ratio and melanosome maturation in melanocytes and melanoma cells.Exp Cell Res. 2001 Aug 1;268(1):26-35. doi: 10.1006/excr.2001.5251. Exp Cell Res. 2001. PMID: 11461115
-
Mahogunin Ring Finger 1 Is Required for Genomic Stability and Modulates the Malignant Phenotype of Melanoma Cells.Cancers (Basel). 2020 Oct 1;12(10):2840. doi: 10.3390/cancers12102840. Cancers (Basel). 2020. PMID: 33019669 Free PMC article.
-
TPC2 controls pigmentation by regulating melanosome pH and size.Proc Natl Acad Sci U S A. 2016 May 17;113(20):5622-7. doi: 10.1073/pnas.1600108113. Epub 2016 May 2. Proc Natl Acad Sci U S A. 2016. PMID: 27140606 Free PMC article.
-
Membrane transport proteins in melanosomes: Regulation of ions for pigmentation.Biochim Biophys Acta Biomembr. 2020 Dec 1;1862(12):183318. doi: 10.1016/j.bbamem.2020.183318. Epub 2020 Apr 22. Biochim Biophys Acta Biomembr. 2020. PMID: 32333855 Free PMC article. Review.
-
Recent advances in understanding the molecular basis of melanogenesis in melanocytes.F1000Res. 2020 Jun 15;9:F1000 Faculty Rev-608. doi: 10.12688/f1000research.24625.1. eCollection 2020. F1000Res. 2020. PMID: 32595944 Free PMC article. Review.
Cited by
-
A Comprehensive Review of Mammalian Pigmentation: Paving the Way for Innovative Hair Colour-Changing Cosmetics.Biology (Basel). 2023 Feb 11;12(2):290. doi: 10.3390/biology12020290. Biology (Basel). 2023. PMID: 36829566 Free PMC article. Review.
-
An MGRN1-Based Biomarker Combination Accurately Predicts Melanoma Patient Survival.Int J Mol Sci. 2025 Feb 18;26(4):1739. doi: 10.3390/ijms26041739. Int J Mol Sci. 2025. PMID: 40004203 Free PMC article.
-
The metabolism of melanin synthesis-From melanocytes to melanoma.Pigment Cell Melanoma Res. 2024 Jul;37(4):438-452. doi: 10.1111/pcmr.13165. Epub 2024 Mar 6. Pigment Cell Melanoma Res. 2024. PMID: 38445351 Free PMC article. Review.
-
Two-pore channel 2 is required for soluble adenylyl cyclase-dependent regulation of melanosomal pH and melanin synthesis.Pigment Cell Melanoma Res. 2024 Sep;37(5):656-666. doi: 10.1111/pcmr.13177. Epub 2024 Jun 6. Pigment Cell Melanoma Res. 2024. PMID: 38844435
-
MGRN1 as a Phenotypic Determinant of Human Melanoma Cells and a Potential Biomarker.Life (Basel). 2022 Jul 26;12(8):1118. doi: 10.3390/life12081118. Life (Basel). 2022. PMID: 35892921 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

