Integrated analysis of DNA methylome and transcriptome reveals the differences in biological characteristics of porcine mesenchymal stem cells
- PMID: 34922435
- PMCID: PMC8684131
- DOI: 10.1186/s12863-021-01016-8
Integrated analysis of DNA methylome and transcriptome reveals the differences in biological characteristics of porcine mesenchymal stem cells
Abstract
Background: Bone marrow (BM) and umbilical cord (UC) are the main sources of mesenchymal stem cells (MSCs). These two MSCs display significant differences in many biological characteristics, yet the underlying regulation mechanisms of these cells remain largely unknown.
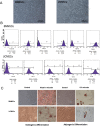
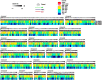
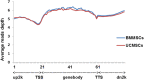
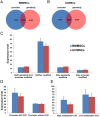
Results: BMMSCs and UCMSCs were isolated from inbred Wuzhishan miniature pigs and the first global DNA methylation and gene expression profiles of porcine MSCs were generated. The osteogenic and adipogenic differentiation ability of porcine BMMSCs is greater than that of UCMSCs. A total of 1979 genes were differentially expressed and 587 genes were differentially methylated at promoter regions in these cells. Integrative analysis revealed that 102 genes displayed differences in both gene expression and promoter methylation. Gene ontology enrichment analysis showed that these genes were associated with cell differentiation, migration, and immunogenicity. Remarkably, skeletal system development-related genes were significantly hypomethylated and upregulated, whereas cell cycle genes were opposite in UCMSCs, implying that these cells have higher cell proliferative activity and lower differentiation potential than BMMSCs.
Conclusions: Our results indicate that DNA methylation plays an important role in regulating the differences in biological characteristics of BMMSCs and UCMSCs. Results of this study provide a molecular theoretical basis for the application of porcine MSCs in human medicine.
Keywords: Bone marrow; DNA methylation; Inbred Wuzhishan miniature pigs; Mesenchymal stem cells; Umbilical cord.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Comparison of transcriptome profiles of mesenchymal stem cells derived from umbilical cord and bone marrow of giant panda (Ailuropoda melanoleuca).Gene. 2022 Dec 15;845:146854. doi: 10.1016/j.gene.2022.146854. Epub 2022 Aug 30. Gene. 2022. PMID: 36055605
-
TNFRSF11B-modified umbilical cord mesenchymal stem cells as a novel strategy for bone-related diseases by suppressing osteoclast activity.J Orthop Surg Res. 2025 May 17;20(1):478. doi: 10.1186/s13018-025-05850-9. J Orthop Surg Res. 2025. PMID: 40380204 Free PMC article.
-
Proteomic analysis of porcine mesenchymal stem cells derived from bone marrow and umbilical cord: implication of the proteins involved in the higher migration capability of bone marrow mesenchymal stem cells.Stem Cell Res Ther. 2015 Apr 15;6(1):77. doi: 10.1186/s13287-015-0061-x. Stem Cell Res Ther. 2015. PMID: 25889491 Free PMC article.
-
Umbilical cord-derived mesenchymal stem cells for hematopoietic stem cell transplantation.J Biomed Biotechnol. 2012;2012:759503. doi: 10.1155/2012/759503. Epub 2012 Oct 3. J Biomed Biotechnol. 2012. PMID: 23093863 Free PMC article. Review.
-
Bone marrow mesenchymal stem cells' osteogenic potential: superiority or non-superiority to other sources of mesenchymal stem cells?Cell Tissue Bank. 2023 Sep;24(3):663-681. doi: 10.1007/s10561-022-10066-w. Epub 2023 Jan 9. Cell Tissue Bank. 2023. PMID: 36622494 Review.
Cited by
-
Epigenetic control of mesenchymal stem cells orchestrates bone regeneration.Front Endocrinol (Lausanne). 2023 Mar 6;14:1126787. doi: 10.3389/fendo.2023.1126787. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36950693 Free PMC article. Review.
-
Homocysteine accelerates hepatocyte autophagy by upregulating TFEB via DNMT3b-mediated DNA hypomethylation.Acta Biochim Biophys Sin (Shanghai). 2023 Apr 6;55(8):1184-1192. doi: 10.3724/abbs.2023060. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37021975 Free PMC article.
-
The Crucial Role of Epigenetic Modifications in Wharton's Jelly Stem Cells.Int J Mol Sci. 2025 Jul 24;26(15):7169. doi: 10.3390/ijms26157169. Int J Mol Sci. 2025. PMID: 40806302 Free PMC article. Review.
-
The Molecular Regulatory Mechanism in Multipotency and Differentiation of Wharton's Jelly Stem Cells.Int J Mol Sci. 2023 Aug 18;24(16):12909. doi: 10.3390/ijms241612909. Int J Mol Sci. 2023. PMID: 37629090 Free PMC article. Review.
References
-
- Shi YF, Hu GZ, Su JJ, Li WZ, Chen Q, Shou PS, Xu CL, Chen XD, Huang Y, Zhu ZX, et al. Mesenchymal stem cells: a new strategy for immunosuppression and tissue repair. Cell Res. 2010;20(5):510–518. - PubMed
-
- Romanov YA, Svintsitskaya VA, Smirnov VN. Searching for alternative sources of postnatal human mesenchymal stem cells: candidate MSC-like cells from umbilical cord. Stem Cells. 2003;21(1):105–110. - PubMed
-
- Semenov OV, Sonja K, Mariluce R, Nikolas Z, Roland Z, Zisch AH, Antoine M. Multipotent mesenchymal stem cells from human placenta: critical parameters for isolation and maintenance of stemness after isolation. Am J Obstet Gynecol. 2010;202(2):193.e191–193.e113. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
