Identifying the hub genes for Duchenne muscular dystrophy and Becker muscular dystrophy by weighted correlation network analysis
- PMID: 34922439
- PMCID: PMC8684282
- DOI: 10.1186/s12863-021-01014-w
Identifying the hub genes for Duchenne muscular dystrophy and Becker muscular dystrophy by weighted correlation network analysis
Abstract
Background: The goal of this study is to identify the hub genes for Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD) via weighted correlation network analysis (WGCNA).
Methods: The gene expression profile of vastus lateralis biopsy samples obtained in 17 patients with DMD, 11 patients with BMD and 6 healthy individuals was downloaded from the Gene Expression Omnibus (GEO) database (GSE109178). After obtaining different expressed genes (DEGs) via GEO2R, WGCNA was conducted using R package, modules and genes that highly associated with DMD, BMD, and their age or pathology were screened. Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment analysis and protein-protein interaction (PPI) network analysis were also conducted. Hub genes and highly correlated clustered genes were identified using Search Tool for the Retrieval of Interacting Genes (STRING) and Cystoscape software.
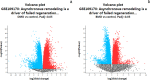
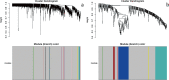
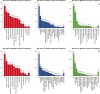
Results: One thousand four hundred seventy DEGs were identified between DMD and control, with 1281 upregulated and 189 downregulated DEGs. Four hundred and twenty DEGs were found between BMD and control, with 157 upregulated and 263 upregulated DEGs. Fourteen modules with different colors were identified for DMD vs control, and 7 modules with different colors were identified for BMD vs control. Ten hub genes were summarized for DMD and BMD respectively, 5 hub genes were summarized for BMD age, 5 and 3 highly correlated clustered genes were summarized for DMD age and BMD pathology, respectively. In addition, 20 GO enrichments were found to be involved in DMD, 3 GO enrichments were found to be involved in BMD, 3 GO enrichments were found to be involved in BMD age.
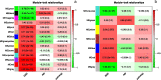
Conclusion: In DMD, several hub genes were identified: C3AR1, TLR7, IRF8, FYB and CD33(immune and inflammation associated genes), TYROBP, PLEK, AIF1(actin reorganization associated genes), LAPTM5 and NT5E(cell death and arterial calcification associated genes, respectively). In BMD, a number of hub genes were identified: LOX, ELN, PLEK, IKZF1, CTSK, THBS2, ADAMTS2, COL5A1(extracellular matrix associated genes), BCL2L1 and CDK2(cell cycle associated genes).
Keywords: Becker muscular dystrophy; Duchenne muscular dystrophy; Gene expression omnibus; Weighted correlation network analysis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Assessment of Weighted Gene Co-Expression Network Analysis to Explore Key Pathways and Novel Biomarkers in Muscular Dystrophy.Pharmgenomics Pers Med. 2021 Apr 13;14:431-444. doi: 10.2147/PGPM.S301098. eCollection 2021. Pharmgenomics Pers Med. 2021. PMID: 33883925 Free PMC article.
-
Identifying hub genes and dysregulated pathways in Duchenne muscular dystrophy.Int J Neurosci. 2025 Apr;135(4):375-387. doi: 10.1080/00207454.2024.2302551. Epub 2024 Jan 17. Int J Neurosci. 2025. PMID: 38179963
-
Identification of hub genes, miRNAs and regulatory factors relevant for Duchenne muscular dystrophy by bioinformatics analysis.Int J Neurosci. 2022 Mar;132(3):296-305. doi: 10.1080/00207454.2020.1810030. Epub 2020 Aug 26. Int J Neurosci. 2022. PMID: 32791870
-
Becker muscular dystrophy: case report, review of the literature, and analysis of differentially expressed hub genes.Neurol Sci. 2022 Jan;43(1):243-253. doi: 10.1007/s10072-021-05499-2. Epub 2021 Nov 3. Neurol Sci. 2022. PMID: 34731335 Review.
-
[From gene to disease; the dystrophin gene involved in Duchenne and Becker muscular dystrophy].Ned Tijdschr Geneeskd. 2002 Feb 23;146(8):364-7. Ned Tijdschr Geneeskd. 2002. PMID: 11887623 Review. Dutch.
Cited by
-
Brothers with Becker muscular dystrophy show discordance in skeletal muscle computed tomography findings: A case report.SAGE Open Med Case Rep. 2024 Jan 3;12:2050313X231221436. doi: 10.1177/2050313X231221436. eCollection 2024. SAGE Open Med Case Rep. 2024. PMID: 38187815 Free PMC article.
-
Anti-fibrotic, muscle-promoting antibody-drug conjugates for the improvement and treatment of DMD.iScience. 2025 Apr 2;28(5):112335. doi: 10.1016/j.isci.2025.112335. eCollection 2025 May 16. iScience. 2025. PMID: 40276765 Free PMC article.
-
CSPG4.CAR-T Cells Modulate Extracellular Matrix Remodeling in DMD Cardiomyopathy.Int J Mol Sci. 2025 Jul 9;26(14):6590. doi: 10.3390/ijms26146590. Int J Mol Sci. 2025. PMID: 40724839 Free PMC article.
-
Application of Droplet Digital PCR Technology in Muscular Dystrophies Research.Int J Mol Sci. 2022 Apr 27;23(9):4802. doi: 10.3390/ijms23094802. Int J Mol Sci. 2022. PMID: 35563191 Free PMC article. Review.
-
Identification of Auxiliary Biomarkers and Description of the Immune Microenvironmental Characteristics in Duchenne Muscular Dystrophy by Bioinformatical Analysis and Experiment.Front Neurosci. 2022 Jun 3;16:891670. doi: 10.3389/fnins.2022.891670. eCollection 2022. Front Neurosci. 2022. PMID: 35720684 Free PMC article.
References
-
- Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82(2):291–329. - PubMed
-
- Emery AE. The muscular dystrophies. Lancet (London, England) 2002;359(9307):687–695. - PubMed
-
- Waldrop MA, Flanigan KM. Update in Duchenne and Becker muscular dystrophy. Curr Opin Neurol. 2019;32(5):722–727. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
