Design and performance of a bovine 200 k SNP chip developed for endangered German Black Pied cattle (DSN)
- PMID: 34922441
- PMCID: PMC8684242
- DOI: 10.1186/s12864-021-08237-2
Design and performance of a bovine 200 k SNP chip developed for endangered German Black Pied cattle (DSN)
Abstract
Background: German Black Pied cattle (DSN) are an endangered dual-purpose breed which was largely replaced by Holstein cattle due to their lower milk yield. DSN cattle are kept as a genetic reserve with a current herd size of around 2500 animals. The ability to track sequence variants specific to DSN could help to support the conservation of DSN's genetic diversity and to provide avenues for genetic improvement.
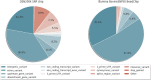
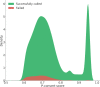
Results: Whole-genome sequencing data of 304 DSN cattle were used to design a customized DSN200k SNP chip harboring 182,154 variants (173,569 SNPs and 8585 indels) based on ten selection categories. We included variants of interest to DSN such as DSN unique variants and variants from previous association studies in DSN, but also variants of general interest such as variants with predicted consequences of high, moderate, or low impact on the transcripts and SNPs from the Illumina BovineSNP50 BeadChip. Further, the selection of variants based on haplotype blocks ensured that the whole-genome was uniformly covered with an average variant distance of 14.4 kb on autosomes. Using 300 DSN and 162 animals from other cattle breeds including Holstein, endangered local cattle populations, and also a Bos indicus breed, performance of the SNP chip was evaluated. Altogether, 171,978 (94.31%) of the variants were successfully called in at least one of the analyzed breeds. In DSN, the number of successfully called variants was 166,563 (91.44%) while 156,684 (86.02%) were segregating at a minor allele frequency > 1%. The concordance rate between technical replicates was 99.83 ± 0.19%.
Conclusion: The DSN200k SNP chip was proved useful for DSN and other Bos taurus as well as one Bos indicus breed. It is suitable for genetic diversity management and marker-assisted selection of DSN animals. Moreover, variants that were segregating in other breeds can be used for the design of breed-specific customized SNP chips. This will be of great value in the application of conservation programs for endangered local populations in the future.
Keywords: Axiom MyDesign; Breed-specific; Conservation; Custom SNP array; Genetic reserve; Holstein cattle.
© 2021. The Author(s).
Conflict of interest statement
All other authors declare that they have no competing interests.
Figures

Similar articles
-
Genetic evaluations for endangered dual-purpose German Black Pied cattle using 50K SNPs, a breed-specific 200K chip, and whole-genome sequencing.J Dairy Sci. 2023 May;106(5):3345-3358. doi: 10.3168/jds.2022-22665. Epub 2023 Apr 5. J Dairy Sci. 2023. PMID: 37028956
-
Genomic diversity and relationship analyses of endangered German Black Pied cattle (DSN) to 68 other taurine breeds based on whole-genome sequencing.Front Genet. 2023 Jan 4;13:993959. doi: 10.3389/fgene.2022.993959. eCollection 2022. Front Genet. 2023. PMID: 36712857 Free PMC article.
-
Whole-Genome Insights into the Genetic Basis of Conformation Traits in German Black Pied (DSN) Cattle.Genes (Basel). 2025 Apr 10;16(4):445. doi: 10.3390/genes16040445. Genes (Basel). 2025. PMID: 40282405 Free PMC article.
-
Genomic Loci Affecting Milk Production in German Black Pied Cattle (DSN).Front Genet. 2021 Mar 8;12:640039. doi: 10.3389/fgene.2021.640039. eCollection 2021. Front Genet. 2021. PMID: 33763120 Free PMC article.
-
Genetic structure and relationships of 16 Asian and European cattle populations using DigiTag2 assay.Anim Sci J. 2016 Feb;87(2):190-6. doi: 10.1111/asj.12416. Epub 2015 Aug 11. Anim Sci J. 2016. PMID: 26260416 Free PMC article. Review.
Cited by
-
Development and application of a cGPS 20K liquid-phase SNP microarray in Jiaji ducks.Poult Sci. 2025 Feb;104(2):104737. doi: 10.1016/j.psj.2024.104737. Epub 2024 Dec 24. Poult Sci. 2025. PMID: 39729728 Free PMC article.
-
Development and validation of a 5K low-density SNP chip for Hainan cattle.BMC Genomics. 2024 Sep 18;25(1):873. doi: 10.1186/s12864-024-10753-w. BMC Genomics. 2024. PMID: 39294563 Free PMC article.
-
Development and verification of a 10K liquid chip for Hainan black goat based on genotyping by pinpoint sequencing of liquid captured targets.BMC Genom Data. 2024 May 7;25(1):44. doi: 10.1186/s12863-024-01228-8. BMC Genom Data. 2024. PMID: 38714950 Free PMC article.
-
Unmapped short reads from whole-genome sequencing indicate potential infectious pathogens in german black Pied cattle.Vet Res. 2023 Oct 18;54(1):95. doi: 10.1186/s13567-023-01227-0. Vet Res. 2023. PMID: 37853447 Free PMC article.
-
Tailoring Genomic Selection for Bos taurus indicus: A Comprehensive Review of SNP Arrays and Reference Genomes.Genes (Basel). 2024 Nov 21;15(12):1495. doi: 10.3390/genes15121495. Genes (Basel). 2024. PMID: 39766762 Free PMC article. Review.
References
-
- Brade W, Brade E. Breeding history of German Holstein cattle. Berichte über Landwirtschaft. 2013;91 https://www.cabdirect.org/cabdirect/abstract/20133389715. Accessed 5 Aug 2019.
-
- GEH e.V. Die Rote Liste im Überblick. The Society for the Conservation of Old and Endangered Livestock Breeds (GEH). 2020. https://www.g-e-h.de/index.php/rote-liste-menu/rote-liste. Accessed 15 Feb 2021.
-
- May K, Weimann C, Scheper C, Strube C, König S. Allele substitution and dominance effects of CD166/ALCAM gene polymorphisms for endoparasite resistance and test-day traits in a small cattle population using logistic regression analyses. Mamm Genome. 2019;30:301–317. doi: 10.1007/s00335-019-09818-z. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

