The versatile role of HuR in Glioblastoma and its potential as a therapeutic target for a multi-pronged attack
- PMID: 34923029
- PMCID: PMC8916685
- DOI: 10.1016/j.addr.2021.114082
The versatile role of HuR in Glioblastoma and its potential as a therapeutic target for a multi-pronged attack
Abstract
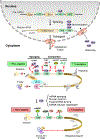
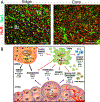
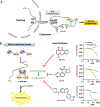
Glioblastoma (GBM) is a malignant and aggressive brain tumor with a median survival of ∼15 months. Resistance to treatment arises from the extensive cellular and molecular heterogeneity in the three major components: glioma tumor cells, glioma stem cells, and tumor-associated microglia and macrophages. Within this triad, there is a complex network of intrinsic and secreted factors that promote classic hallmarks of cancer, including angiogenesis, resistance to cell death, proliferation, and immune evasion. A regulatory node connecting these diverse pathways is at the posttranscriptional level as mRNAs encoding many of the key drivers contain adenine- and uridine rich elements (ARE) in the 3' untranslated region. Human antigen R (HuR) binds to ARE-bearing mRNAs and is a major positive regulator at this level. This review focuses on basic concepts of ARE-mediated RNA regulation and how targeting HuR with small molecule inhibitors represents a plausible strategy for a multi-pronged therapeutic attack on GBM.
Keywords: Adenine- and Uridine-rich elements; Cytoplasmic translocation; GBM; HuR multimerization; Posttranscriptional regulation; Small molecule inhibitors.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

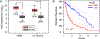
References
-
- Tamimi AF, Juweid M, Epidemiology and Outcome of Glioblastoma, in: De Vleeschouwer S (Ed.) Glioblastoma, Brisbane (AU), 2017. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

