Hepatic TM6SF2 Is Required for Lipidation of VLDL in a Pre-Golgi Compartment in Mice and Rats
- PMID: 34923175
- PMCID: PMC8804273
- DOI: 10.1016/j.jcmgh.2021.12.008
Hepatic TM6SF2 Is Required for Lipidation of VLDL in a Pre-Golgi Compartment in Mice and Rats
Abstract
Background & aims: Substitution of lysine for glutamic acid at residu 167 in Transmembrane 6 superfamily member 2 (TM6SF2) is associated with fatty liver disease and reduced plasma lipid levels. Tm6sf2-/- mice replicate the human phenotype but were not suitable for detailed mechanistic studies. As an alternative model, we generated Tm6sf2-/- rats to determine the subcellular location and function of TM6SF2.
Methods: Two lines of Tm6sf2-/- rats were established using gene editing. Lipids from tissues and from newly secreted very low density lipoproteins (VLDLs) were quantified using enzymatic assays and mass spectrometry. Neutral lipids were visualized in tissue sections using Oil Red O staining. The rate of dietary triglyceride (TG) absorption and hepatic VLDL-TG secretion were compared in Tm6sf2-/- mice and in their wild-type littermates. The intracellular location of TM6SF2 was determined by cell fractionation. Finally, TM6SF2 was immunoprecipitated from liver and enterocytes to identify interacting proteins.
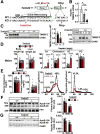
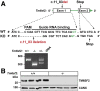
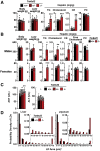
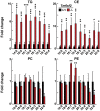
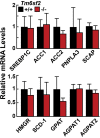
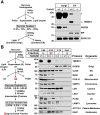
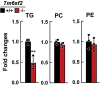
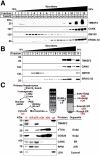
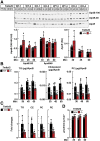
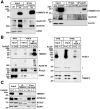
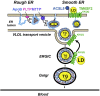
Results: Tm6sf2-/- rats had a 6-fold higher mean hepatic TG content (56.1 ± 28.9 9 vs 9.8 ± 3.9 mg/g; P < .0001) and lower plasma cholesterol levels (99.0 ± 10.5 vs 110.6 ± 14.0 mg/dL; P = .0294) than their wild-type littermates. Rates of appearance of dietary and hepatic TG into blood were reduced significantly in Tm6sf2-/- rats (P < .001 and P < .01, respectively). Lipid content of newly secreted VLDLs isolated from perfused livers was reduced by 53% (TG) and 62% (cholesterol) (P = .005 and P = .01, respectively) in Tm6sf2-/- mice. TM6SF2 was present predominantly in the smooth endoplasmic reticulum and endoplasmic reticulum-Golgi intermediate compartments, but not in Golgi. Both apolipoprotein B-48 and acyl-CoA synthetase long chain family member 5 physically interacted with TM6SF2.
Conclusions: TM6SF2 acts in the smooth endoplasmic reticulum to promote bulk lipidation of apolipoprotein B-containing lipoproteins, thus preventing fatty liver disease.
Keywords: Fatty Liver; Liver Perfusion; Triglycerides; VLDL.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. - PubMed
-
- Buch S., Stickel F., Trepo E., Way M., Herrmann A., Nischalke H.D., Brosch M., Rosendahl J., Berg T., Ridinger M., Rietschel M., McQuillin A., Frank J., Kiefer F., Schreiber S., Lieb W., Soyka M., Semmo N., Aigner E., Datz C., Schmelz R., Bruckner S., Zeissig S., Stephan A.M., Wodarz N., Deviere J., Clumeck N., Sarrazin C., Lammert F., Gustot T., Deltenre P., Volzke H., Lerch M.M., Mayerle J., Eyer F., Schafmayer C., Cichon S., Nothen M.M., Nothnagel M., Ellinghaus D., Huse K., Franke A., Zopf S., Hellerbrand C., Moreno C., Franchimont D., Morgan M.Y., Hampe J. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

