Autophagy modulates cell fate decisions during lineage commitment
- PMID: 34923909
- PMCID: PMC9450964
- DOI: 10.1080/15548627.2021.2008691
Autophagy modulates cell fate decisions during lineage commitment
Abstract
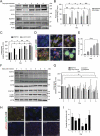
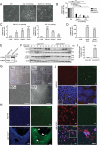
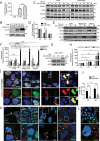
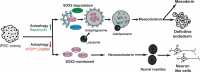
Early events during development leading to exit from a pluripotent state and commitment toward a specific germ layer still need in-depth understanding. Autophagy has been shown to play a crucial role in both development and differentiation. This study employs human embryonic and induced pluripotent stem cells to understand the early events of lineage commitment with respect to the role of autophagy in this process. Our data indicate that a dip in autophagy facilitates exit from pluripotency. Upon exit, we demonstrate that the modulation of autophagy affects SOX2 levels and lineage commitment, with induction of autophagy promoting SOX2 degradation and mesendoderm formation, whereas inhibition of autophagy causes SOX2 accumulation and neuroectoderm formation. Thus, our results indicate that autophagy-mediated SOX2 turnover is a determining factor for lineage commitment. These findings will deepen our understanding of development and lead to improved methods to derive different lineages and cell types.Abbreviations: ACTB: Actin, beta; ATG: Autophagy-related; BafA1: Bafilomycin A1; CAS9: CRISPR-associated protein 9; CQ: Chloroquine; DE: Definitive endoderm; hESCs: Human Embryonic Stem Cells; hiPSCs: Human Induced Pluripotent Stem Cells; LAMP1: Lysosomal Associated Membrane Protein 1; MAP1LC3: Microtubule-Associated Protein 1 Light Chain 3; MTOR: Mechanistic Target Of Rapamycin Kinase; NANOG: Nanog Homeobox; PAX6: Paired Box 6; PE: Phosphatidylethanolamine; POU5F1: POU class 5 Homeobox 1; PRKAA2: Protein Kinase AMP-Activated Catalytic Subunit Alpha 2; SOX2: SRY-box Transcription Factor 2; SQSTM1: Sequestosome 1; ULK1: unc-51 like Autophagy Activating Kinase 1; WDFY3: WD Repeat and FYVE Domain Containing 3.
Keywords: Autophagosome; SOX2; differentiation; ectoderm; endoderm; mesoderm; pluripotent stem cells.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
