TALE-induced cell death executors: an origin outside immunity?
- PMID: 34924289
- PMCID: PMC7612725
- DOI: 10.1016/j.tplants.2021.11.003
TALE-induced cell death executors: an origin outside immunity?
Abstract
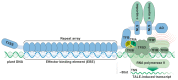
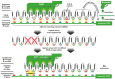
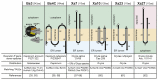
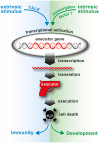
Phytopathogenic bacteria inject effector proteins into plant host cells to promote disease. Plant resistance (R) genes encoding nucleotide-binding leucine-rich repeat (NLR) proteins mediate the recognition of functionally and structurally diverse microbial effectors, including transcription-activator like effectors (TALEs) from the bacterial genus Xanthomonas. TALEs bind to plant promoters and transcriptionally activate either disease-promoting host susceptibility (S) genes or cell death-inducing executor-type R genes. It is perplexing that plants contain TALE-perceiving executor-type R genes in addition to NLRs that also mediate the recognition of TALE-containing xanthomonads. We present recent findings on the evolvability of TALEs, which suggest that the native function of executors is not in plant immunity, but possibly in the regulation of developmentally controlled programmed cell death (PCD) processes.
Keywords: Xanthomonas; evolution; executor; nucleotide-binding leucine-rich repeat (NLR) protein; programmed cell death (PCD); transcription activator-like effector (TALE).
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- de Lange O, et al. From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J. 2014;78:753–771. - PubMed
-
- Pérez-Quintero AL, Szurek B. A decade decoded: spies and hackers in the history of TAL effectors research. Annu Rev Phytopathol. 2019;57:459–481. - PubMed
-
- Becker S, Boch J. TALE and TALEN genome editing technologies. Gene and Genome Editing. 2021;2:100007
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

