Nicotinic receptors as SARS-CoV-2 spike co-receptors?
- PMID: 34924680
- PMCID: PMC8669939
- DOI: 10.1016/j.mehy.2021.110741
Nicotinic receptors as SARS-CoV-2 spike co-receptors?
Abstract
Nicotinic acetylcholine receptors (nAChRs) play an important role in homeostasis and respiratory diseases. Controversies regarding the association between COVID-19 hospitalizations and smoking suggest that nAChRs may contribute to SARS-CoV-2 respiratory syndrome. We recently detailed the expression and localization of all nAChR subunits in the human lung. Since virus association with nAChRs has been shown in the past, we hypothesize that nAChR subunits act as SARS-CoV-2 Spike co-receptors. Based on sequence alignment analysis, we report domains of high molecular similarities in nAChRs with the binding domain of hACE2 for SARS-CoV-2 Spike protein. This hypothesis supported by in silico pilot data provides a rational for the modelling and the in vitro experimental validation of the interaction between SARS-CoV-2 and the nAChRs.
Keywords: ACE-2, Angiotensin converting enzyme-2; BR, Binding region; COVID-19; COVID-19, Coronavirus disease 2019; ETD, Extracellular topological domain; Lung; Nicotinic receptors; PDB, Protein data bank; RBD, Receptor binding domain; SARS-CoV-2; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; hACE2; nAChRs, Nicotinic acetylcholine receptors; vdw, Van der Waals.
© 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
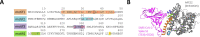

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

