Repurposing of phytomedicine-derived bioactive compounds with promising anti-SARS-CoV-2 potential: Molecular docking, MD simulation and drug-likeness/ADMET studies
- PMID: 34924801
- PMCID: PMC8667520
- DOI: 10.1016/j.sjbs.2021.12.018
Repurposing of phytomedicine-derived bioactive compounds with promising anti-SARS-CoV-2 potential: Molecular docking, MD simulation and drug-likeness/ADMET studies
Abstract
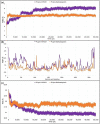
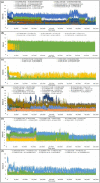
In view of the potential of traditional plant-based remedies (or phytomedicines) in the management of COVID-19, the present investigation was aimed at finding novel anti-SARS-CoV-2 molecules by in silico screening of bioactive phytochemicals (database) using computational methods and drug repurposing approach. A total of 160 compounds belonging to various phytochemical classes (flavonoids, limonoids, saponins, triterpenoids, steroids etc.) were selected (as initial hits) and screened against three specific therapeutic targets (Mpro/3CLpro, PLpro and RdRp) of SARS-CoV-2 by docking, molecular dynamics simulation and drug-likeness/ADMET studies. From our studies, six phytochemicals were identified as notable ant-SARS-CoV-2 agents (best hit molecules) with promising inhibitory effects effective against protease (Mpro and PLpro) and polymerase (RdRp) enzymes. These compounds are namely, ginsenoside Rg2, saikosaponin A, somniferine, betulinic acid, soyasapogenol C and azadirachtin A. On the basis of binding modes and dynamics studies of protein-ligand intercations, ginsenoside Rg2, saikosaponin A, somniferine were found to be the most potent (in silico) inhibitors potentially active against Mpro, PLpro and RdRp, respectively. The present investigation can be directed towards further experimental studies in order to confirm the anti-SARS-CoV-2 efficacy along with toxicities of identified phytomolecules.
Keywords: Drug repurposing; Molecular docking; Molecular dynamics; Phytochemicals; Phytomedicine; SARS-CoV-2 infection.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
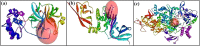
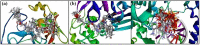




References
-
- Adeoye A.O., Oso B.J., Olaoye I.F., Tijjani H., Adebayo A.I. Repurposing of chloroquine and some clinically approved antiviral drugs aseffective therapeutics to prevent cellular entry and replication of coronavirus. J. Biomol. Struct. Dyn. 2020;1–14 doi: 10.1080/07391102.2020.1765876. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

