Multimodal Label-Free Monitoring of Adipogenic Stem Cell Differentiation Using Endogenous Optical Biomarkers
- PMID: 34924914
- PMCID: PMC8680429
- DOI: 10.1002/adfm.202103955
Multimodal Label-Free Monitoring of Adipogenic Stem Cell Differentiation Using Endogenous Optical Biomarkers
Abstract
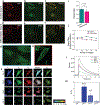
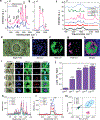
Stem cell-based therapies carry significant promise for treating human diseases. However, clinical translation of stem cell transplants for effective treatment requires precise non-destructive evaluation of the purity of stem cells with high sensitivity (<0.001% of the number of cells). Here, a novel methodology using hyperspectral imaging (HSI) combined with spectral angle mapping-based machine learning analysis is reported to distinguish differentiating human adipose-derived stem cells (hASCs) from control stem cells. The spectral signature of adipogenesis generated by the HSI method enables identifying differentiated cells at single-cell resolution. The label-free HSI method is compared with the standard techniques such as Oil Red O staining, fluorescence microscopy, and qPCR that are routinely used to evaluate adipogenic differentiation of hASCs. HSI is successfully used to assess the abundance of adipocytes derived from transplanted cells in a transgenic mice model. Further, Raman microscopy and multiphoton-based metabolic imaging is performed to provide complementary information for the functional imaging of the hASCs. Finally, the HSI method is validated using matrix-assisted laser desorption/ionization-mass spectrometry imaging of the stem cells. The study presented here demonstrates that multimodal imaging methods enable label-free identification of stem cell differentiation with high spatial and chemical resolution.
Keywords: MALDI; Raman microscopy; hyperspectral imaging; label-free stem cell imaging; second harmonic generation.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures
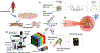




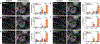


References
-
- Caplan AI, J. Orthop. Res 1991, 9, 641. - PubMed
-
- Baksh D, Yao R, Tuan RS, Stem Cells 2007, 25, 1384. - PubMed
-
- Gimble JM, Guilak F, Cytotherapy 2003, 5, 362. - PubMed
-
- a) Wu L, Wang T, Ge Y, Cai X, Wang J, Lin Y, Cell Proliferation 2012, 45, 311; - PMC - PubMed
- b) Greenberg AS, Obin MS, Am. J. Clin. Nutr 2006, 83, 461S; - PubMed
- c) Fantuzzi G, J. Allergy Clin. Immunol 2005, 115, 911; - PubMed
- d) Shaik S, Wu X, Gimble J, Devireddy R, Sci. Rep 2018, 8, 8162. - PMC - PubMed
-
- Suhito IR, Han Y, Min J, Son H, Kim T-H, Biomaterials 2018, 154, 223. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
