Genomic Analysis of Global Staphylococcus argenteus Strains Reveals Distinct Lineages With Differing Virulence and Antibiotic Resistance Gene Content
- PMID: 34925305
- PMCID: PMC8677677
- DOI: 10.3389/fmicb.2021.795173
Genomic Analysis of Global Staphylococcus argenteus Strains Reveals Distinct Lineages With Differing Virulence and Antibiotic Resistance Gene Content
Abstract
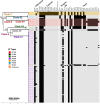
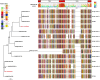
Infections due to Staphylococcus argenteus have been increasingly reported worldwide and the microbe cannot be distinguished from Staphylococcus aureus by standard methods. Its complement of virulence determinants and antibiotic resistance genes remain unclear, and how far these are distinct from those produced by S. aureus remains undetermined. In order to address these uncertainties, we have collected 132 publicly available sequences from fourteen different countries, including the United Kingdom, between 2005 and 2018 to study the global genetic structure of the population. We have compared the genomes for antibiotic resistance genes, virulence determinants and mobile genetic elements such as phages, pathogenicity islands and presence of plasmid groups between different clades. 20% (n = 26) isolates were methicillin resistant harboring a mecA gene and 88% were penicillin resistant, harboring the blaZ gene. ST2250 was identified as the most frequent strain, but ST1223, which was the second largest group, contained a marginally larger number of virulence genes compared to the other STs. Novel S. argenteus pathogenicity islands were identified in our isolates harboring tsst-1, seb, sec3, ear, selk, selq toxin genes, as well as chromosomal clusters of enterotoxin and superantigen-like genes. Strain-specific type I modification systems were widespread which would limit interstrain transfer of genetic material. In addition, ST2250 possessed a CRISPR/Cas system, lacking in most other STs. S. argenteus possesses important genetic differences from S. aureus, as well as between different STs, with the potential to produce distinct clinical manifestations.
Keywords: CRISPR/Cas; Staphylococcus argenteus; Staphylococcus argenteus pathogenicity islands; antibiotic resistance; virulence genes.
Copyright © 2021 Goswami, Fox, Holden, Leanord and Evans.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Argudín M. A., Dodémont M., Vandendriessche S., Rottiers S., Tribes C., Roisin S., et al. (2016). Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Europ. J. Clin. Microbiol. Infect. Dis. 35 1017–1022. 10.1007/s10096-016-2632-x - DOI - PubMed
LinkOut - more resources
Full Text Sources

