Comprehensive in Vitro Characterization of the LSD1 Small Molecule Inhibitor Class in Oncology
- PMID: 34927013
- PMCID: PMC8669716
- DOI: 10.1021/acsptsci.1c00223
Comprehensive in Vitro Characterization of the LSD1 Small Molecule Inhibitor Class in Oncology
Abstract
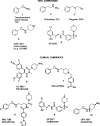
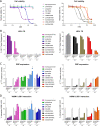
Lysine-specific demethylase 1 (LSD1 or KDM1A) is a chromatin modifying enzyme playing a key role in the cell cycle and cell differentiation and proliferation through the demethylation of histones and nonhistone substrates. In addition to its enzymatic activity, LSD1 plays a fundamental scaffolding role as part of transcription silencing complexes such as rest co-repressor (CoREST) and nucleosome remodeling and deacetylase (NuRD). A host of classical amine oxidase inhibitors such as tranylcypromine, pargyline, and phenelzine together with LSD1 tool compounds such as SP-2509 and GSK-LSD1 have been extensively utilized in LSD1 mechanistic cancer studies. Additionally, several optimized new chemical entities have reached clinical trials in oncology such as ORY-1001 (iadademstat), GSK2879552, SP-2577 (seclidemstat), IMG-7289 (bomedemstat), INCB059872, and CC-90011 (pulrodemstat). Despite this, no single study exists that characterizes them all under the same experimental conditions, preventing a clear interpretation of published results. Herein, we characterize the whole LSD1 small molecule compound class as inhibitors of LSD1 catalytic activity, disruptors of SNAIL/GFI1 (SNAG)-scaffolding protein-protein interactions, inducers of cell differentiation, and potential anticancer treatments for hematological and solid tumors to yield an updated, unified perspective of this field. Our results highlight significant differences in potency and selectivity among the clinical compounds with iadademstat being the most potent and reveal that most of the tool compounds have very low activity and selectivity, suggesting some conclusions derived from their use should be taken with caution.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Verma M.; Kumar V.. Epigenetic Drugs for Cancer and Precision Medicine. In Epigenetics of Aging and Longevity; Elsevier, 2018; pp 439–451;10.1016/B978-0-12-811060-7.00021-8. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
