Chromodomain helicase DNA-binding 4 (CHD4) regulates early B cell identity and V(D)J recombination
- PMID: 34927255
- PMCID: PMC9351717
- DOI: 10.1111/imr.13054
Chromodomain helicase DNA-binding 4 (CHD4) regulates early B cell identity and V(D)J recombination
Abstract
B lymphocytes develop from uncommitted precursors into immunoglobulin (antibody)-producing B cells, a major arm of adaptive immunity. Progression of early progenitors to antibody-expressing cells in the bone marrow is orchestrated by the temporal regulation of different gene programs at discrete developmental stages. A major question concerns how B cells control the accessibility of these genes to transcription factors. Research has implicated nucleosome remodeling ATPases as mediators of chromatin accessibility. Here, we describe studies of chromodomain helicase DNA-binding 4 (CHD4; also known as Mi-2β) in early B cell development. CHD4 comprises multiple domains that function in nucleosome mobilization and histone binding. CHD4 is a key component of Nucleosome Remodeling and Deacetylase, or NuRD (Mi-2) complexes, which assemble with other proteins that mediate transcriptional repression. We review data demonstrating that CHD4 is necessary for B lineage identity: early B lineage progression, proliferation in response to interleukin-7, responses to DNA damage, and cell survival in vivo. CHD4-NuRD is also required for the Ig heavy-chain repertoire by promoting utilization of distal variable (VH ) gene segments in V(D)J recombination. In conclusion, the regulation of chromatin accessibility by CHD4 is essential for production of antibodies by B cells, which in turn mediate humoral immune responses to pathogens and disease.
Keywords: B cell identity; CHD4; V(D)J recombination; chromodomain helicase DNA-binding 4.
© 2021 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures
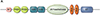
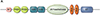


References
-
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature.1998;395:917–921. - PubMed
-
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molec Cell.1998;2:851–861. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

